3I8O
 
 | | A domain of a functionally unknown protein from Methanocaldococcus jannaschii DSM 2661. | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, K, Chhor, G, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-09 | | Release date: | 2009-07-21 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.638 Å) | | Cite: | A domain of a functionally unknown protein from Methanocaldococcus jannaschii DSM 2661.
To be Published
|
|
3IVP
 
 | |
3IUV
 
 | | The structure of a member of TetR family (SCO1917) from Streptomyces coelicolor A3 | | Descriptor: | uncharacterized TetR family protein | | Authors: | Tan, K, Cuff, M, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | The structure of a member of TetR family (SCO1917) from Streptomyces coelicolor A3
To be Published
|
|
3K6H
 
 | | Crystal structure of a nitroreductase family protein from Agrobacterium tumefaciens str. C58 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Nitroreductase family protein, SULFATE ION | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-08 | | Release date: | 2009-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of a nitroreductase family protein from Agrobacterium tumefaciens str. C58
To be Published
|
|
3KKC
 
 | | The crystal structure OF TetR transcriptional regulator from Streptococcus agalactiae 2603V | | Descriptor: | IMIDAZOLE, NICKEL (II) ION, TetR family Transcriptional regulator | | Authors: | Tan, K, Hatzos, C, Morgan, T, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-11-05 | | Release date: | 2009-11-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure OF TetR transcriptional regulator from Streptococcus agalactiae 2603V
To be Published
|
|
3L1W
 
 | | The crystal structure of a functionally unknown conserved protein from Enterococcus faecalis V583 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, SULFATE ION, ... | | Authors: | Tan, K, Rakowski, E, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of a functionally unknown conserved protein from Enterococcus faecalis V583
To be Published
|
|
7XXF
 
 | | Structure of photosynthetic LH1-RC super-complex of Rhodopila globiformis | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (6~{E},8~{E},10~{E},12~{E},14~{E},16~{E},18~{E},20~{E},22~{E},24~{E},26~{E},28~{E})-2,31-dimethoxy-2,6,10,14,19,23,27,31-octamethyl-dotriaconta-6,8,10,12,14,16,18,20,22,24,26,28-dodecaen-5-one, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Tani, K, Kanno, R, Kurosawa, K, Takaichi, S, Nagashima, K.V.P, Hall, M, Yu, L.-J, Kimura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Wang-Otomo, Z.-Y. | | Deposit date: | 2022-05-30 | | Release date: | 2022-11-16 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | An LH1-RC photocomplex from an extremophilic phototroph provides insight into origins of two photosynthesis proteins.
Commun Biol, 5, 2022
|
|
8CZQ
 
 | | The crystal structure of MtbTOP1 in complex with both G- and T-segments | | Descriptor: | ACETATE ION, DNA (5'-D(*CP*TP*TP*CP*CP*GP*CP*TP*TP*GP*AP*C)-3'), DNA topoisomerase 1, ... | | Authors: | Tan, K, Tse-Dinh, Y.-C. | | Deposit date: | 2022-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The interaction between transport-segment DNA and topoisomerase IA-crystal structure of MtbTOP1 in complex with both G- and T-segments.
Nucleic Acids Res., 51, 2023
|
|
8FWF
 
 | | Crystal structure of Apo form Fab235 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Tan, K, Kim, M, Reinherz, E.L. | | Deposit date: | 2023-01-21 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
8FXJ
 
 | | Crystal structure of Fab460 | | Descriptor: | ACETATE ION, CHLORIDE ION, Fab460, ... | | Authors: | Tan, K, Kim, M, Reinherz, E.L. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
8FYM
 
 | | Crystal structure of Fab235 in complex with MPER peptide | | Descriptor: | ALA-SER-LEU-TRP-ASN-TRP-PHE-ASN-ILE-THR-ASN-TRP-LEU-TRP-TYR-ILE-LYS-LYS-LYS, CHLORIDE ION, Fab235, ... | | Authors: | Tan, K, Kim, M, Reinherz, E.L. | | Deposit date: | 2023-01-26 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
8FZ2
 
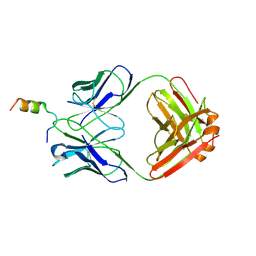 | | Crystal structure of Fab460 in complex with MPER peptide | | Descriptor: | Fab460, H chain, L chain, ... | | Authors: | Tan, K, Kim, M, Reinherz, E.L. | | Deposit date: | 2023-01-27 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
7N6H
 
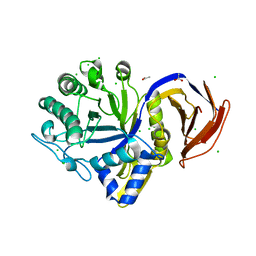 | |
7N6O
 
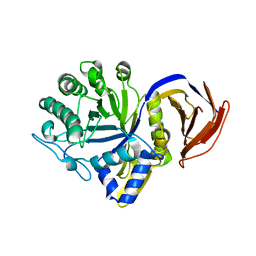 | |
7TVX
 
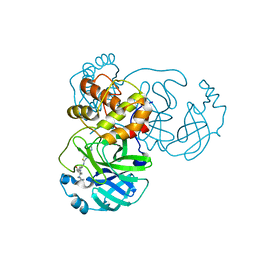 | |
7TVS
 
 | | The Crystal Structure of SARS-CoV-2 Omicron Mpro (P132H) in complex with demethylated analog of masitinib | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N-(4-methyl-3-{[4-(pyridin-3-yl)-1,3-thiazol-2-yl]amino}phenyl)-4-[(piperazin-1-yl)methyl]benzamide | | Authors: | Tan, K, Maltseva, N.I, Endres, M.J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-02-05 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88612878 Å) | | Cite: | The Crystal Structure of SARS-CoV-2 Omicron Mpro (P132H) in complex with demethylated analog of masitinib
To Be Published
|
|
7TYE
 
 | |
5IZN
 
 | | The crystal structure of 50S ribosomal protein L25 from Vibrio vulnificus CMCP6 | | Descriptor: | 50S ribosomal protein L25, PHOSPHATE ION | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-03-25 | | Release date: | 2016-04-06 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of 50S ribosomal protein L25 from Vibrio vulnificus CMCP6
To Be Published
|
|
5JRO
 
 | | The crystal structure of azoreductase from Yersinia pestis CO92 in its Apo form | | Descriptor: | FMN-dependent NADH-azoreductase, GLYCEROL | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-06 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | The crystal structure of azoreductase from Yersinia pestis CO92 in its Apo form
To Be Published
|
|
8TFG
 
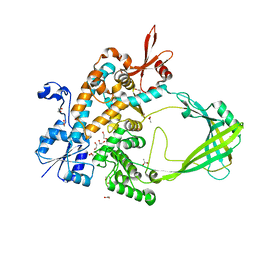 | |
6E4B
 
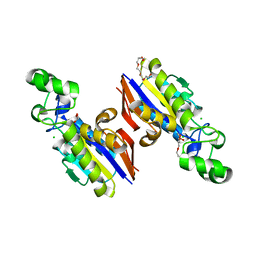 | |
5JMU
 
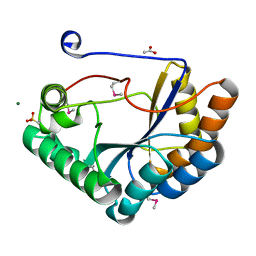 | | The crystal structure of the catalytic domain of peptidoglycan N-acetylglucosamine deacetylase from Eubacterium rectale ATCC 33656 | | Descriptor: | ACETATE ION, MAGNESIUM ION, Peptidoglycan N-acetylglucosamine deacetylase, ... | | Authors: | Tan, K, Gu, M, Clancy, S, Joachimiak, A. | | Deposit date: | 2016-04-29 | | Release date: | 2016-06-29 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The crystal structure of the catalytic domain of peptidoglycan N-acetylglucosamine deacetylase from Eubacterium rectale ATCC 33656 (CASP target)
To Be Published
|
|
5JMB
 
 | |
3LDU
 
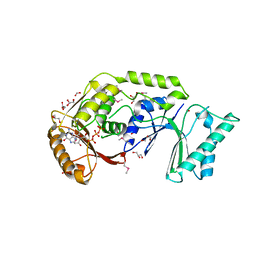 | | The crystal structure of a possible methylase from Clostridium difficile 630. | | Descriptor: | FORMIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Tan, K, Wu, R, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-13 | | Release date: | 2010-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a possible methylase from Clostridium difficile 630.
To be Published
|
|
3M1R
 
 | | The crystal structure of formimidoylglutamase from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | CACODYLATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tan, K, Bigelow, L, Trevino, D, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-05 | | Release date: | 2010-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | The crystal structure of formimidoylglutamase from Bacillus subtilis subsp. subtilis str. 168
To be Published
|
|
