1IOF
 
 | | X-RAY CRYSTALLINE STRUCTURES OF PYRROLIDONE CARBOXYL PEPTIDASE FROM A HYPERTHERMOPHILE, PYROCOCCUS FURIOSUS, AND ITS CYS-FREE MUTANT | | Descriptor: | PYRROLIDONE CARBOXYL PEPTIDASE | | Authors: | Tanaka, H, Chinami, M, Ota, M, Tsukihara, T, Yutani, K. | | Deposit date: | 2001-03-09 | | Release date: | 2001-03-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystalline structures of pyrrolidone carboxyl peptidase from a hyperthermophile, Pyrococcus furiosus, and its cys-free mutant.
J.Biochem., 130, 2001
|
|
7WAG
 
 | | Crystal structure of MurJ squeezed form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DI(HYDROXYETHYL)ETHER, Lipid II flippase MurJ | | Authors: | Tsukazaki, T, Kohga, H, Tanaka, Y, Yoshikaie, K, Taniguchi, K, Fujimoto, K. | | Deposit date: | 2021-12-14 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the lipid flippase MurJ in a "squeezed" form distinct from its inward- and outward-facing forms.
Structure, 30, 2022
|
|
4EJ7
 
 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, ATP-bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Aminoglycoside 3'-phosphotransferase AphA1-IAB, CALCIUM ION, ... | | Authors: | Stogios, P.J, Minasov, G, Tan, K, Evdokimova, E, Egorova, O, Di Leo, R, Shakya, T, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-06 | | Release date: | 2012-04-18 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
7WAW
 
 | | MurJ inward closed form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, lipid II flippase MurJ | | Authors: | Tsukazaki, T, Kohga, H, Tanaka, Y, Yoshikaie, K, Taniguchi, K, Fujimoto, K. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the lipid flippase MurJ in a "squeezed" form distinct from its inward- and outward-facing forms.
Structure, 30, 2022
|
|
7WAX
 
 | | MurJ inward occluded form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (4S)-2-METHYL-2,4-PENTANEDIOL, lipid II flippase MurJ | | Authors: | Tsukazaki, T, Kohga, H, Tanaka, Y, Yoshikaie, K, Taniguchi, K, Fujimoto, K. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the lipid flippase MurJ in a "squeezed" form distinct from its inward- and outward-facing forms.
Structure, 30, 2022
|
|
4FC9
 
 | | Structure of the C-terminal domain of the type III effector Xcv3220 (XopL) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, uncharacterized protein | | Authors: | Singer, A.U, Xu, X, Cui, H, Tan, K, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-24 | | Release date: | 2012-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the C-terminal domain of the type III effector Xcv3220 (XopL)
To be Published
|
|
7WSS
 
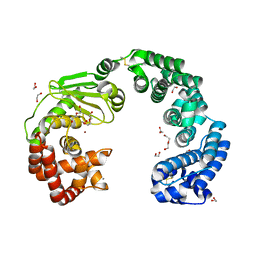 | | Collagenase from Grimontia (Vibrio) hollisae 1706B | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ikeuchi, T, Yasumoto, M, Takita, T, Mizutani, K, Mikami, B, Tanaka, K, Hattori, S, Yasukawa, K. | | Deposit date: | 2022-02-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of Grimontia hollisae collagenase provides insights into its novel substrate specificity toward collagen.
J.Biol.Chem., 298, 2022
|
|
7XEB
 
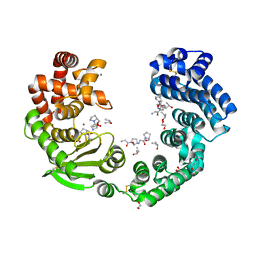 | | Collagenase from Grimontia (Vibrio) hollisae 1706B complexed with Gly-Pro-Hyp | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLY-PRO-HYP peptide, ... | | Authors: | Ikeuchi, T, Yasumoto, M, Takita, T, Mizutani, K, Mikami, B, Tanaka, K, Hattori, S, Yasukawa, K. | | Deposit date: | 2022-03-30 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of Grimontia hollisae collagenase provides insights into its novel substrate specificity toward collagen.
J.Biol.Chem., 298, 2022
|
|
1TAB
 
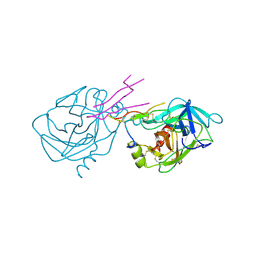 | | STRUCTURE OF THE TRYPSIN-BINDING DOMAIN OF BOWMAN-BIRK TYPE PROTEASE INHIBITOR AND ITS INTERACTION WITH TRYPSIN | | Descriptor: | BOWMAN-BIRK TYPE PROTEINASE INHIBITOR, TRYPSIN | | Authors: | Tsunogae, Y, Tanaka, I, Yamane, T, Kikkawa, J.-I, Ashida, T, Ishikawa, C, Watanabe, K, Nakamura, S, Takahashi, K. | | Deposit date: | 1990-10-15 | | Release date: | 1992-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the trypsin-binding domain of Bowman-Birk type protease inhibitor and its interaction with trypsin.
J.Biochem.(Tokyo), 100, 1986
|
|
4DE4
 
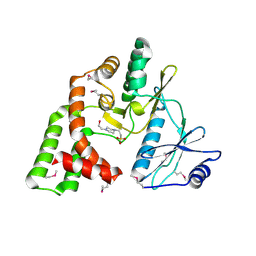 | | Crystal structure of aminoglycoside phosphotransferase APH(2")-Id/APH(2")-IVa in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, APH(2")-Id | | Authors: | Stogios, P.J, Minasov, G, Tan, K, Nocek, B, Evdokimova, E, Egorova, O, Di Leo, R, Li, H, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-19 | | Release date: | 2012-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A small molecule discrimination map of the antibiotic resistance kinome.
Chem.Biol., 18, 2011
|
|
4XRE
 
 | | Crystal structure of Gnk2 complexed with mannose | | Descriptor: | Antifungal protein ginkbilobin-2, alpha-D-mannopyranose | | Authors: | Miyakawa, T, Hatano, K, Miyauchi, Y, Suwa, Y, Sawano, Y, Tanokura, M. | | Deposit date: | 2015-01-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | A secreted protein with plant-specific cysteine-rich motif functions as a mannose-binding lectin that exhibits antifungal activity.
Plant Physiol., 166, 2014
|
|
2D1I
 
 | | Structure of human Atg4b | | Descriptor: | Cysteine protease APG4B | | Authors: | Kumanomidou, T, Mizushima, T, Komatsu, M, Suzuki, A, Tanida, I, Sou, Y.S, Ueno, T, Kominami, E, Tanaka, K, Yamane, T. | | Deposit date: | 2005-08-24 | | Release date: | 2006-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human Atg4b, a Processing and De-conjugating Enzyme for Autophagosome-forming Modifiers
J.Mol.Biol., 355, 2006
|
|
2D1V
 
 | | Crystal structure of DNA-binding domain of Bacillus subtilis YycF | | Descriptor: | Transcriptional regulatory protein yycF | | Authors: | Okajima, T, Okada, A, Watanabe, T, Yamamoto, K, Tanizawa, K, Utsumi, R. | | Deposit date: | 2005-09-01 | | Release date: | 2006-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Response regulator YycF essential for bacterial growth: X-ray crystal structure of the DNA-binding domain and its PhoB-like DNA recognition motif
Febs Lett., 582, 2008
|
|
3A1M
 
 | | A fusion protein of a beta helix region of gene product 5 and the foldon region of bacteriophage T4 | | Descriptor: | POTASSIUM ION, chimera of thrombin cleavage site, Tail-associated lysozyme, ... | | Authors: | Yokoi, N, Suzuki, A, Hikage, T, Koshiyama, T, Terauchi, M, Yutani, K, Kanamaru, S, Arisaka, F, Yamane, T, Watanabe, Y, Ueno, T. | | Deposit date: | 2009-04-11 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Construction of Robust Bio-nanotubes using the Controlled Self-Assembly of Component Proteins of Bacteriophage T4
Small, 6, 2010
|
|
4V8X
 
 | | Structure of Thermus thermophilus ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Feng, S, Chen, Y, Kamada, K, Wang, H, Tang, K, Wang, M, Gao, Y.G. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Yoeb-Ribosome Structure: A Canonical Rnase that Requires the Ribosome for its Specific Activity.
Nucleic Acids Res., 41, 2013
|
|
2ZWB
 
 | | Neutron crystal structure of wild type human lysozyme in D2O | | Descriptor: | Lysozyme C | | Authors: | Chiba-Kamoshida, K, Matsui, T, Chatake, T, Ohhara, T, Ostermann, A, Tanaka, I, Yutani, K, Niimura, N. | | Deposit date: | 2008-12-02 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-16 | | Method: | NEUTRON DIFFRACTION (1.8 Å) | | Cite: | Site-specific softening of peptide bonds by localized deuterium observed by neutron crystallography of human lysozyme
To be Published
|
|
3AA9
 
 | | Crystal Structure Analysis of the Mutant CutA1 (E61V) from E. coli | | Descriptor: | Divalent-cation tolerance protein cutA | | Authors: | Matsuura, Y, Tanaka, T, Bagautdinov, B, Kunishima, N, Yutani, K. | | Deposit date: | 2009-11-12 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Remarkable improvement in the heat stability of CutA1 from Escherichia coli by rational protein design
J.Biochem., 148, 2010
|
|
7F8O
 
 | | Cryo-EM structure of the C-terminal deletion mutant of human PANX1 in a nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7F8J
 
 | | Cryo-EM structure of human pannexin-1 in a nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7F8N
 
 | | Human pannexin-1 showing a conformational change in the N-terminal domain and blocked pore | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
3A8Q
 
 | | Low-resolution crystal structure of the Tiam2 PHCCEx domain | | Descriptor: | T-lymphoma invasion and metastasis-inducing protein 2 | | Authors: | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
3A8P
 
 | | Crystal structure of the Tiam2 PHCCEx domain | | Descriptor: | T-lymphoma invasion and metastasis-inducing protein 2 | | Authors: | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
3A8N
 
 | | Crystal structure of the Tiam1 PHCCEx domain | | Descriptor: | T-lymphoma invasion and metastasis-inducing protein 1 | | Authors: | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
3AH6
 
 | | Remarkable improvement of the heat stability of CutA1 from E.coli by rational protein designing | | Descriptor: | Divalent-cation tolerance protein cutA | | Authors: | Matsuura, Y, Tanaka, T, Bagautdinov, B, Kunishima, N, Yutani, K. | | Deposit date: | 2010-04-15 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Remarkable improvement in the heat stability of CutA1 from Escherichia coli by rational protein design
J.Biochem., 148, 2010
|
|
3AA8
 
 | | Crystal Structure Analysis of the Mutant CutA1 (S11V/E61V) from E. coli | | Descriptor: | Divalent-cation tolerance protein cutA | | Authors: | Matsuura, Y, Tanaka, T, Bagautdinov, B, Kunishima, N, Yutani, K. | | Deposit date: | 2009-11-12 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Remarkable improvement in the heat stability of CutA1 from Escherichia coli by rational protein design
J.Biochem., 148, 2010
|
|
