3A77
 
 | | The crystal structure of phosphorylated IRF-3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Interferon regulatory factor 3 | | Authors: | Takahasi, K, Horiuchi, M, Noda, N.N, Inagaki, F. | | Deposit date: | 2009-09-17 | | Release date: | 2010-08-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ser386 phosphorylation of transcription factor IRF-3 induces dimerization and association with CBP/p300 without overall conformational change.
Genes Cells, 15, 2010
|
|
1J2F
 
 | | X-ray crystal structure of IRF-3 and its functional implications | | Descriptor: | Interferon regulatory factor 3 | | Authors: | Takahasi, K, Noda, N, Horiuchi, M, Mori, M, Okabe, Y, Fukuhara, Y, Terasawa, H, Fujita, T, Inagaki, F. | | Deposit date: | 2003-01-04 | | Release date: | 2003-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of IRF-3 and its functional implications.
Nat.Struct.Biol., 10, 2003
|
|
2RQE
 
 | | Solution structure of the silkworm bGRP/GNBP3 N-terminal domain reveals the mechanism for b-1,3-glucan specific recognition | | Descriptor: | Beta-1,3-glucan-binding protein | | Authors: | Takahasi, K, Ochiai, M, Horiuchi, M, Kumeta, H, Ogura, K, Ashida, M, Inagaki, F. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the silkworm betaGRP/GNBP3 N-terminal domain reveals the mechanism for beta-1,3-glucan-specific recognition.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2RQA
 
 | | Solution structure of LGP2 CTD | | Descriptor: | ATP-dependent RNA helicase DHX58, ZINC ION | | Authors: | Takahasi, K, Kumeta, H, Tsuduki, N, Narita, R, Shigemoto, T, Hirai, R, Yoneyama, M, Horiuchi, M, Ogura, K, Fujita, T, Fuyuhiko, I. | | Deposit date: | 2009-03-17 | | Release date: | 2009-05-05 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Cytosolic RNA Sensor MDA5 and LGP2 C-terminal Domains: IDENTIFICATION OF THE RNA RECOGNITION LOOP IN RIG-I-LIKE RECEPTORS
J.Biol.Chem., 284, 2009
|
|
2RQB
 
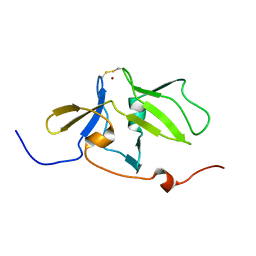 | | Solution structure of MDA5 CTD | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, ZINC ION | | Authors: | Takahasi, K, Kumeta, H, Tsuduki, N, Narita, R, Shigemoto, T, Hirai, R, Yoneyama, M, Horiuchi, M, Ogura, K, Fujita, T, Fuyuhiko, I. | | Deposit date: | 2009-03-17 | | Release date: | 2009-05-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Cytosolic RNA Sensor MDA5 and LGP2 C-terminal Domains: IDENTIFICATION OF THE RNA RECOGNITION LOOP IN RIG-I-LIKE RECEPTORS
J.Biol.Chem., 284, 2009
|
|
2RMJ
 
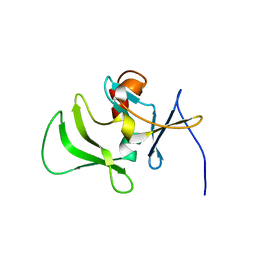 | | Solution structure of RIG-I C-terminal domain | | Descriptor: | Probable ATP-dependent RNA helicase DDX58 | | Authors: | Takahasi, K, Yoneyama, M, Nihishori, T, Hirai, R, Narita, R, Gale Jr, M, Fujita, T, Inagaki, F. | | Deposit date: | 2007-10-23 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nonself RNA-Sensing Mechanism of RIG-I Helicase and Activation of Antiviral Immune Responses
Mol.Cell, 29, 2008
|
|
2D5R
 
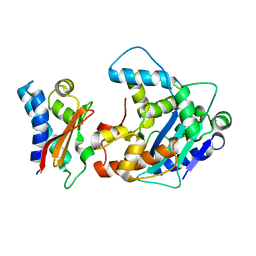 | | Crystal Structure of a Tob-hCaf1 Complex | | Descriptor: | CCR4-NOT transcription complex subunit 7, Tob1 protein | | Authors: | Horiuchi, M, Suzuki, N.N, Muroya, N, Takahasi, K, Nishida, M, Yoshida, Y, Ikematsu, N, Nakamura, T, Kawamura-Tsuzuku, J, Yamamoto, T, Inagaki, F. | | Deposit date: | 2005-11-04 | | Release date: | 2006-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the antiproliferative activity of the Tob-hCaf1 complex.
J.Biol.Chem., 284, 2009
|
|
