6P7J
 
 | |
6P50
 
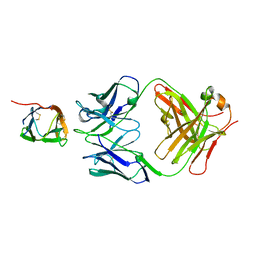 | | Crystal Structure of a Complex of human IL-7Ralpha with an anti-IL-7Ralpha Fab 4A10 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-7 receptor subunit alpha, anti-IL-7R 4A10 Fab heavy chain, ... | | 著者 | Walsh, S.T.R, Kashi, L, Kohnhorst, C.L. | | 登録日 | 2019-05-29 | | 公開日 | 2019-09-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | New anti-IL-7R alpha monoclonal antibodies show efficacy against T cell acute lymphoblastic leukemia in pre-clinical models.
Leukemia, 34, 2020
|
|
6UIZ
 
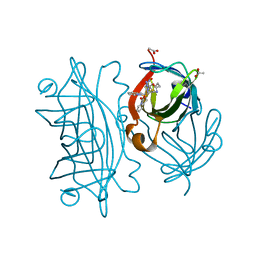 | | Artificial Iron Proteins: Modelling the Active Sites in Non-Heme Dioxygenases | | 分子名称: | ACETATE ION, Streptavidin, {N-(2-{bis[(pyridin-2-yl-kappaN)methyl]amino-kappaN}ethyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}(triaza-1,2-dien-2-ium-1-ide-kappaN~1~)iron(4+) | | 著者 | Miller, K.R, Paretsky, J.D, Follmer, A.H, Heinisch, T, Mittra, K, Gul, S, Kim, I.-S, Fuller, F.D, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bhowmick, A, Sauter, N.K, Kern, J, Yano, J, Green, M.T, Ward, T.R, Borovik, A.S. | | 登録日 | 2019-10-01 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Artificial Iron Proteins: Modeling the Active Sites in Non-Heme Dioxygenases.
Inorg.Chem., 59, 2020
|
|
4AUK
 
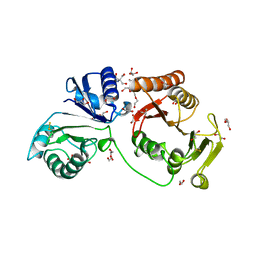 | | Crystal structure of C2498 2'-O-ribose methyltransferase RlmM from Escherichia coli | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Punekar, A.S, Shepherd, T.R, Liljeruhm, J, Forster, A.C, Selmer, M. | | 登録日 | 2012-05-18 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of Rlmm, the 2'O-Ribose Methyltransferase for C2498 of Escherichia Coli 23S Rrna.
Nucleic Acids Res., 40, 2012
|
|
6OI9
 
 | | Crystal Structure of E. coli Biotin Carboxylase Complexed with 7-[3-(aminomethyl)pyrrolidin-1-yl]-6-(2,6-dichlorophenyl)pyrido[2,3-d]pyrimidin-2-amine | | 分子名称: | 1,2-ETHANEDIOL, 7-[(3S)-3-(aminomethyl)pyrrolidin-1-yl]-6-(2,6-dichlorophenyl)pyrido[2,3-d]pyrimidin-2-amine, Biotin carboxylase | | 著者 | Andrews, L.D, Kane, T.R, Dozzo, P, Haglund, C.M, Hilderbrandt, D.J, Linsell, M.S, Machajewski, T, McEnroe, G, Serio, A.W, Wlasichuk, K.B, Neau, D.B, Pakhomova, S, Waldrop, G.L, Sharp, M, Pogliano, J, Cirz, R, Cohen, F. | | 登録日 | 2019-04-09 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Optimization and Mechanistic Characterization of Pyridopyrimidine Inhibitors of Bacterial Biotin Carboxylase.
J.Med.Chem., 62, 2019
|
|
4ATO
 
 | | New insights into the mechanism of bacterial Type III toxin-antitoxin systems: selective toxin inhibition by a non-coding RNA pseudoknot | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, TOXI, TOXN | | 著者 | Short, F.L, Pei, X.Y, Blower, T.R, Ong, S.L, Luisi, B.F, Salmond, G.P.C. | | 登録日 | 2012-05-09 | | 公開日 | 2012-12-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Selectivity and Self-Assembly in the Control of a Bacterial Toxin by an Antitoxic Noncoding RNA Pseudoknot.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4C1O
 
 | | Geobacillus thermoglucosidasius GH family 52 xylosidase | | 分子名称: | 1,2-ETHANEDIOL, BETA-XYLOSIDASE, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Espina, G, Eley, K, Schneider, T.R, Crennell, S.J, Danson, M.J. | | 登録日 | 2013-08-13 | | 公開日 | 2014-05-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A Novel Beta-Xylosidase Structure from Geobacillus Thermoglucosidasius: The First Crystal Structure of a Glycoside Hydrolase Family Gh52 Enzyme Reveals Unpredicted Similarity to Other Glycoside Hydrolase Folds
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1VIB
 
 | | NMR SOLUTION STRUCTURE OF THE NEUROTOXIN B-IV, 20 STRUCTURES | | 分子名称: | NEUROTOXIN B-IV | | 著者 | Barnham, K.J, Dyke, T.R, Kem, W.R, Norton, R.S. | | 登録日 | 1996-11-25 | | 公開日 | 1997-05-15 | | 最終更新日 | 2017-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of neurotoxin B-IV from the marine worm Cerebratulus lacteus: a helical hairpin cross-linked by disulphide bonding.
J.Mol.Biol., 268, 1997
|
|
1WLG
 
 | | Crystal structure of FlgE31, a major fragment of the hook protein | | 分子名称: | Flagellar hook protein flgE | | 著者 | Samatey, F.A, Matsunami, H, Imada, K, Nagashima, S, Shaikh, T.R, Thomas, D.R, DeRosier, D.J, Kitao, A, Namba, K. | | 登録日 | 2004-06-25 | | 公開日 | 2004-11-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of the bacterial flagellar hook and implication for the molecular universal joint mechanism.
Nature, 431, 2004
|
|
6UI0
 
 | | Artificial Iron Proteins: Modelling the Active Sites in Non-Heme Dioxygenases | | 分子名称: | ACETATE ION, Streptavidin, {N-(2-{bis[(pyridin-2-yl-kappaN)methyl]amino-kappaN}ethyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}iron(3+) | | 著者 | Miller, K.R, Paretsky, J.D, Follmer, A.H, Heinisch, T, Mittra, K, Gul, S, Kim, I.-S, Fuller, F.D, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bhowmick, A, Sauter, N.K, Kern, J, Yano, J, Green, M.T, Ward, T.R, Borovik, A.S. | | 登録日 | 2019-09-29 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Artificial Iron Proteins: Modeling the Active Sites in Non-Heme Dioxygenases.
Inorg.Chem., 59, 2020
|
|
1W7Z
 
 | | Crystal structure of the free (uncomplexed) Ecballium elaterium trypsin inhibitor (EETI-II) | | 分子名称: | FORMIC ACID, SODIUM ION, TRYPSIN INHIBITOR II | | 著者 | Kraetzner, R, Debreczeni, J.E, Pape, T, Kolmar, H, Schneider, T.R, Uson, I, Scheldrick, G.M. | | 登録日 | 2004-09-14 | | 公開日 | 2005-11-01 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Structure of Ecballium Elaterium Trypsin Inhibitor II (Eeti-II): A Rigid Molecular Scaffold
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4ATN
 
 | | Crystal structure of C2498 2'-O-ribose methyltransferase RlmM from Escherichia coli | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, RIBOSOMAL RNA LARGE SUBUNIT METHYLTRANSFERASE M, ... | | 著者 | Punekar, A.S, Shepherd, T.R, Liljeruhm, J, Forster, A.C, Selmer, M. | | 登録日 | 2012-05-08 | | 公開日 | 2012-08-15 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal Structure of Rlmm, the 2'O-Ribose Methyltransferase for C2498 of Escherichia Coli 23S Rrna.
Nucleic Acids Res., 40, 2012
|
|
4AX1
 
 | | Q157N mutant. Crystal Structure of the Mobile Metallo-beta-Lactamase AIM-1 from Pseudomonas aeruginosa: Insights into Antibiotic Binding and the role of Gln157 | | 分子名称: | ACETATE ION, CALCIUM ION, MAGNESIUM ION, ... | | 著者 | Leiros, H.-K.S, Borra, P.S, Brandsdal, B.O, Edvardsen, K.S.W, Spencer, J, Walsh, T.R, Samuelsen, O. | | 登録日 | 2012-06-06 | | 公開日 | 2012-06-20 | | 最終更新日 | 2019-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal Structure of the Mobile Metallo-Beta-Lactamase Aim-1 from Pseudomonas Aeruginosa: Insights Into Antibiotic Binding and the Role of Gln157
Antimicrob.Agents Chemother., 56, 2012
|
|
1US0
 
 | | Human Aldose Reductase in complex with NADP+ and the inhibitor IDD594 at 0.66 Angstrom | | 分子名称: | ALDOSE REDUCTASE, CITRIC ACID, IDD594, ... | | 著者 | Howard, E.I, Sanishvili, R, Cachau, R.E, Mitschler, A, Chevrier, B, Barth, P, Lamour, V, Van Zandt, M, Sibley, E, Bon, C, Moras, D, Schneider, T.R, Joachimiak, A, Podjarny, A. | | 登録日 | 2003-11-16 | | 公開日 | 2004-05-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (0.66 Å) | | 主引用文献 | Ultrahigh Resolution Drug Design I: Details of Interactions in Human Aldose Reductase-Inhibitor Complex at 0.66 A.
Proteins, 55, 2004
|
|
1NBQ
 
 | | Crystal Structure of Human Junctional Adhesion Molecule Type 1 | | 分子名称: | Junctional adhesion molecule 1 | | 著者 | Prota, A.E, Campbell, J.A, Schelling, P, Forrest, J.C, Watson, M.J, Peters, T.R, Aurrand-Lions, M, Imhof, B.A, Dermody, T.S, Stehle, T. | | 登録日 | 2002-12-03 | | 公開日 | 2003-04-01 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of human junctional adhesion molecule 1: Implications for reovirus binding
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1X0N
 
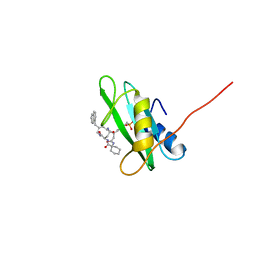 | | NMR structure of growth factor receptor binding protein SH2 domain complexed with the inhibitor | | 分子名称: | 4-[(10S,14S,18S)-18-(2-AMINO-2-OXOETHYL)-14-(1-NAPHTHYLMETHYL)-8,17,20-TRIOXO-7,16,19-TRIAZASPIRO[5.14]ICOS-11-EN-10-YL]BENZYLPHOSPHONIC ACID, Growth factor receptor-bound protein 2 | | 著者 | Ogura, K, Shiga, T, Yuzawa, S, Yokochi, M, Burke, T.R, Inagaki, F. | | 登録日 | 2005-03-24 | | 公開日 | 2005-04-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of growth factor receptor binding protein SH2 domain complexed with the inhibitor
To be Published
|
|
8C24
 
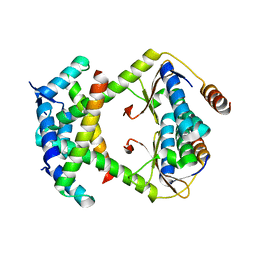 | |
8C26
 
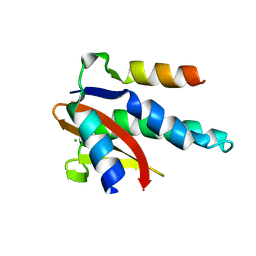 | |
4BLG
 
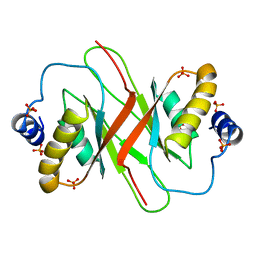 | | Crystal structure of MHV-68 Latency-associated nuclear antigen (LANA) C-terminal DNA binding domain | | 分子名称: | LATENCY-ASSOCIATED NUCLEAR ANTIGEN, PHOSPHATE ION | | 著者 | Correia, B, Cerqueira, S.A, Beauchemin, C, Pires De Miranda, M, Li, S, Ponnusamy, R, Rodrigues, L, Schneider, T.R, Carrondo, M.A, Kaye, K.M, Simas, J.P, McVey, C.E. | | 登録日 | 2013-05-02 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of the Gamma-2 Herpesvirus Lana DNA Binding Domain Identifies Charged Surface Residues which Impact Viral Latency
Plos Pathog., 9, 2013
|
|
1UPD
 
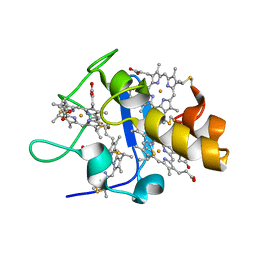 | | Oxidized STRUCTURE OF CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 AT PH 7.6 | | 分子名称: | CYTOCHROME C3, HEME C | | 著者 | Bento, I, Matias, P.M, Baptista, A.M, Da Costa, P.N, Van Dongen, W.M.A.M, Saraiva, L.M, Schneider, T.R, Soares, C.M, Carrondo, M.A. | | 登録日 | 2003-09-29 | | 公開日 | 2004-09-30 | | 最終更新日 | 2019-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Molecular Basis for Redox-Bohr and Cooperative Effects in Cytochrome C3 from Desulfovibrio Desulfuricans Atcc 27774: Crystallographic and Modeling Studies of Oxidized and Reduced High-Resolution Structures at Ph 7.6
Proteins, 54, 2004
|
|
6UOG
 
 | |
6VE1
 
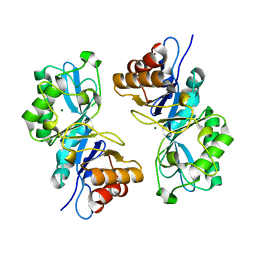 | |
6UOD
 
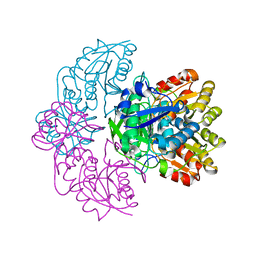 | |
1UP9
 
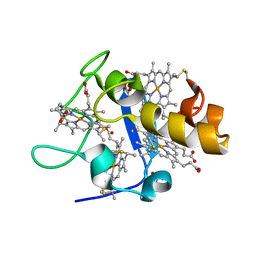 | | REDUCED STRUCTURE OF CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 AT PH 7.6 | | 分子名称: | CYTOCHROME C3, HEME C, SULFATE ION | | 著者 | Bento, I, Matias, P.M, Baptista, A.M, Da Costa, P.N, Van Dongen, W.M.A.M, Saraiva, L.M, Schneider, T.R, Soares, C.M, Carrondo, M.A. | | 登録日 | 2003-09-29 | | 公開日 | 2004-09-30 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Molecular Basis for Redox-Bohr and Cooperative Effects in Cytochrome C3 from Desulfovibrio Desulfuricans Atcc 27774: Crystallographic and Modeling Studies of Oxidized and Reduced High-Resolution Structures at Ph 7.6
Proteins, 54, 2004
|
|
4C1P
 
 | | Geobacillus thermoglucosidasius GH family 52 xylosidase | | 分子名称: | BETA-XYLOSIDASE, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | 著者 | Espina, G, Eley, K, Schneider, T.R, Crennell, S.J, Danson, M.J. | | 登録日 | 2013-08-13 | | 公開日 | 2014-05-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.634 Å) | | 主引用文献 | A Novel Beta-Xylosidase Structure from Geobacillus Thermoglucosidasius: The First Crystal Structure of a Glycoside Hydrolase Family Gh52 Enzyme Reveals Unpredicted Similarity to Other Glycoside Hydrolase Folds
Acta Crystallogr.,Sect.D, 70, 2014
|
|
