6NOC
 
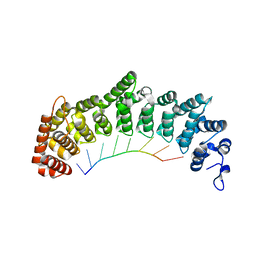 | | Crystal structure of FBF-2 repeat 5 mutant (C363A, R364Y) in complex with 8-nt RNA | | Descriptor: | Fem-3 mRNA-binding factor 2, RNA (5'-R(*UP*GP*UP*AP*AP*AP*UP*A)-3') | | Authors: | McCann, K, Wang, Y, Qiu, C, Hall, T.M.T. | | Deposit date: | 2019-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.849 Å) | | Cite: | Engineering a conserved RNA regulatory protein repurposes its biological function in vivo .
Elife, 8, 2019
|
|
6NDJ
 
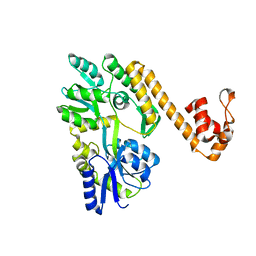 | | Crystal structure of human NLRP6 PYD domain with MBP fusion | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein, NACHT, LRR and PYD domains-containing protein 6 chimera | | Authors: | Shen, C, Fu, T.M, Wu, H. | | Deposit date: | 2018-12-13 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Molecular mechanism for NLRP6 inflammasome assembly and activation.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3QC5
 
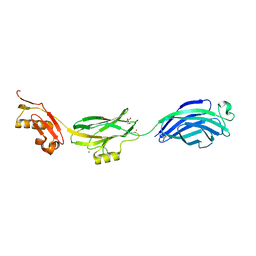 | | GspB | | Descriptor: | GLYCEROL, NITROGEN MOLECULE, POTASSIUM ION, ... | | Authors: | Pyburn, T.M, Iverson, T.M. | | Deposit date: | 2011-01-15 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Structural Model for Binding of the Serine-Rich Repeat Adhesin GspB to Host Carbohydrate Receptors.
Plos Pathog., 7, 2011
|
|
6NMW
 
 | |
6NOH
 
 | | Crystal structure of FBF-2 repeat 5 mutant (C363S, R364Y, Q367S) in complex with 8-nt RNA | | Descriptor: | 1,2-ETHANEDIOL, Fem-3 mRNA-binding factor 2, RNA (5'-R(*UP*GP*UP*AP*AP*AP*UP*A)-3') | | Authors: | McCann, K.L, Wang, Y, Qiu, C, Hall, T.M.T. | | Deposit date: | 2019-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Engineering a conserved RNA regulatory protein repurposes its biological function in vivo .
Elife, 8, 2019
|
|
4GYY
 
 | | Crystal structure of human O-GlcNAc Transferase with UDP-5SGlcNAc and a peptide substrate | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Lazarus, M.B, Jiang, J, Gloster, T.M, Zandberg, W.F, Vocadlo, D.J, Walker, S. | | Deposit date: | 2012-09-05 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural snapshots of the reaction coordinate for O-GlcNAc transferase.
Nat.Chem.Biol., 8, 2012
|
|
6NCV
 
 | | Cryo-EM structure of NLRP6 PYD filament | | Descriptor: | NACHT, LRR and PYD domains-containing protein 6 | | Authors: | Shen, C, Fu, T.M, Wu, H. | | Deposit date: | 2018-12-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular mechanism for NLRP6 inflammasome assembly and activation.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5T38
 
 | | Crystal Structure of the N-terminal domain of EvdMO1 with SAH bound | | Descriptor: | EvdMO1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McCulloch, K.M, Berndt, S, Yamakawa, I, Chen, Q, Loukachevitch, L.V, Starbird, C, Perry, N.A, Iverson, T.M. | | Deposit date: | 2016-08-25 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1502 Å) | | Cite: | The Structure of the Bifunctional Everninomicin Biosynthetic Enzyme EvdMO1 Suggests Independent Activity of the Fused Methyltransferase-Oxidase Domains.
Biochemistry, 57, 2018
|
|
1DVW
 
 | | NMR structure of 18 residue peptide from merp protein | | Descriptor: | 18 RESIDUE PEPTIDE FROM MERP PROTEIN, MERCURY (II) ION | | Authors: | Veglia, G, Porcelli, F, De Silva, T.M, Prantner, A.M, Opella, S.J. | | Deposit date: | 2000-01-22 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Metal-Binding Motif
GMTCAAC Is Similar in an 18-Residue Linear
Peptide and the Mercury Binding Protein MerP
J.Am.Chem.Soc., 122, 2000
|
|
6MG0
 
 | | Crystal structure of a 5-domain construct of LgrA in the thiolation state | | Descriptor: | 5'-({[(2R,3R)-3-amino-2-{[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-{[oxido(oxo)phosphonio]oxy}butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}-4-methylpentyl]sulfonyl}amino)-5'-deoxyadenosine, Linear gramicidin synthase subunit A | | Authors: | Reimer, J.M, Eivaskhani, M, Harb, I, Schmeing, T.M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility.
Science, 366, 2019
|
|
1DOK
 
 | | MONOCYTE CHEMOATTRACTANT PROTEIN 1, P-FORM | | Descriptor: | MONOCYTE CHEMOATTRACTANT PROTEIN 1, SULFATE ION | | Authors: | Lubkowski, J, Bujacz, G, Boque, L, Wlodawer, A, Domaille, P.J, Handel, T.M. | | Deposit date: | 1996-11-27 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of MCP-1 in two crystal forms provides a rare example of variable quaternary interactions.
Nat.Struct.Biol., 4, 1997
|
|
5UZG
 
 | | Crystal structure of Glorund qRRM1 domain | | Descriptor: | AT27789p, GLYCEROL, SULFATE ION | | Authors: | Teramoto, T, Hall, T.M.T. | | Deposit date: | 2017-02-26 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | The Drosophila hnRNP F/H Homolog Glorund Uses Two Distinct RNA-Binding Modes to Diversify Target Recognition.
Cell Rep, 19, 2017
|
|
5V5D
 
 | | Room temperature (280K) crystal structure of Kaposi's sarcoma-associated herpesvirus protease in complex with allosteric inhibitor (compound 250) | | Descriptor: | 4-{[6-(cyclohexylmethyl)pyridine-2-carbonyl]amino}-3-(phenylamino)benzoic acid, ORF 17 | | Authors: | Thompson, M.C, Acker, T.M, Fraser, J.S, Craik, C.S. | | Deposit date: | 2017-03-14 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Allosteric Inhibitors, Crystallography, and Comparative Analysis Reveal Network of Coordinated Movement across Human Herpesvirus Proteases.
J. Am. Chem. Soc., 139, 2017
|
|
6NBS
 
 | | WT ERK2 with compound 2507-8 | | Descriptor: | (5S)-5-benzyl-4,5-dihydro-1H-imidazol-2-amine, GLYCEROL, Mitogen-activated protein kinase 1, ... | | Authors: | Sammons, R.M, Perry, N.A, Cho, E.J, Kaoud, T.S, Zamora-Olivares, D.P, Piserchio, A, Houghten, R.A, Giulianotti, M, Li, Y, Debevec, G, Gurevich, V.V, Ghose, R, Iverson, T.M, Dalby, K.N. | | Deposit date: | 2018-12-10 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Novel Class of Common Docking Domain Inhibitors That Prevent ERK2 Activation and Substrate Phosphorylation.
Acs Chem.Biol., 14, 2019
|
|
6NWW
 
 | |
1FUR
 
 | |
6NYV
 
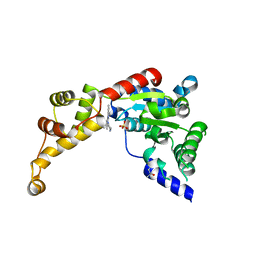 | | Structure of spastin AAA domain in complex with a quinazoline-based inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, N-(3-tert-butyl-1H-pyrazol-5-yl)-2-[(2R)-2-methylpiperazin-1-yl]quinazolin-4-amine, SULFATE ION, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-02-12 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.425 Å) | | Cite: | Designing Allele-Specific Inhibitors of Spastin, a Microtubule-Severing AAA Protein.
J. Am. Chem. Soc., 141, 2019
|
|
5UTE
 
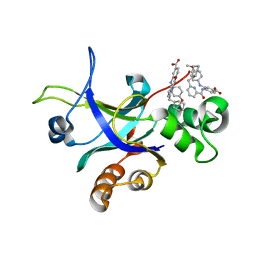 | |
1EK5
 
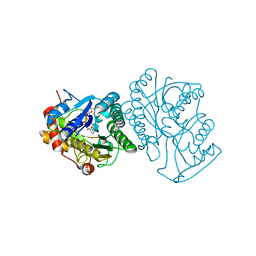 | | STRUCTURE OF HUMAN UDP-GALACTOSE 4-EPIMERASE IN COMPLEX WITH NAD+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-GALACTOSE 4-EPIMERASE | | Authors: | Thoden, J.B, Wohlers, T.M, Fridovich-Keil, J.L, Holden, H.M. | | Deposit date: | 2000-03-06 | | Release date: | 2000-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic evidence for Tyr 157 functioning as the active site base in human UDP-galactose 4-epimerase.
Biochemistry, 39, 2000
|
|
6NZC
 
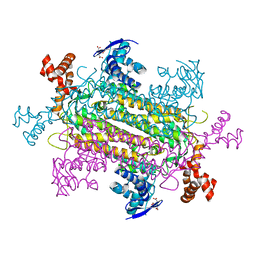 | |
4GZ5
 
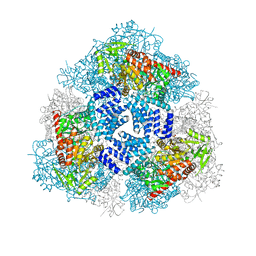 | | Crystal structure of human O-GlcNAc Transferase with UDP-GlcNAc | | Descriptor: | SULFATE ION, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Lazarus, M.B, Jiang, J, Gloster, T.M, Zandberg, W.F, Vocadlo, D.J, Walker, S. | | Deposit date: | 2012-09-06 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.075 Å) | | Cite: | Structural snapshots of the reaction coordinate for O-GlcNAc transferase.
Nat.Chem.Biol., 8, 2012
|
|
6NOF
 
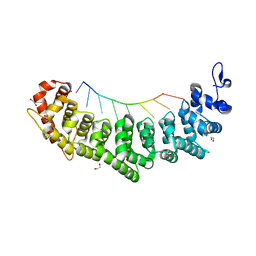 | | Crystal structure of FBF-2 repeat 5 mutant (C363A, R364Y, Q367S) in complex with 8-nt RNA | | Descriptor: | 1,2-ETHANEDIOL, Fem-3 mRNA-binding factor 2, RNA (5'-R(*UP*GP*UP*AP*AP*AP*UP*A)-3') | | Authors: | McCann, K.L, Wang, Y, Qiu, C, Hall, T.M.T. | | Deposit date: | 2019-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Engineering a conserved RNA regulatory protein repurposes its biological function in vivo .
Elife, 8, 2019
|
|
4GYW
 
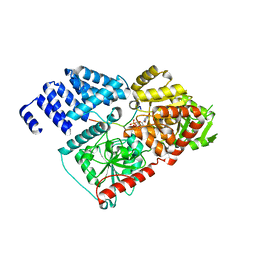 | | Crystal structure of human O-GlcNAc Transferase in complex with UDP and a glycopeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Lazarus, M.B, Jiang, J, Gloster, T.M, Zandberg, W.F, Vocadlo, D.J, Walker, S. | | Deposit date: | 2012-09-05 | | Release date: | 2012-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural snapshots of the reaction coordinate for O-GlcNAc transferase.
Nat.Chem.Biol., 8, 2012
|
|
6NUI
 
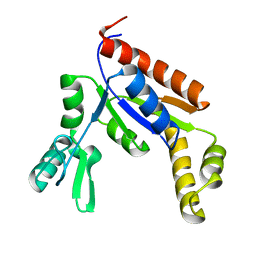 | | Human Guanylate Kinase | | Descriptor: | Guanylate kinase | | Authors: | Sabo, T.M, Khan, N, Ban, D, Trigo-Mourino, P, Carneiro, M.G, Trent, J.O, Konrad, M, Lee, D. | | Deposit date: | 2019-02-01 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and functional investigation of human guanylate kinase reveals allosteric networking and a crucial role for the enzyme in cancer.
J.Biol.Chem., 294, 2019
|
|
6NZ9
 
 | | Crystal structure of E. coli fumarase C bound to citrate at 1.53 angstrom resolution | | Descriptor: | CITRIC ACID, Fumarate hydratase class II | | Authors: | Stuttgen, G.M, May, J.F, Bhattcharyya, B, Weaver, T.M. | | Deposit date: | 2019-02-13 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.528 Å) | | Cite: | Closed fumarase C active-site structures reveal SS Loop residue contribution in catalysis.
Febs Lett., 594, 2020
|
|
