6P4J
 
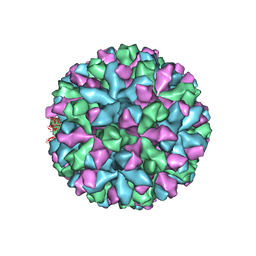 | | Mouse norovirus complexed with GCDCA | | Descriptor: | Capsid protein, GLYCOCHENODEOXYCHOLIC ACID | | Authors: | Smith, T.J, Smith, T.J. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Bile Salts Alter the Mouse Norovirus Capsid Conformation: Possible Implications for Cell Attachment and Immune Evasion.
J.Virol., 93, 2019
|
|
3L6V
 
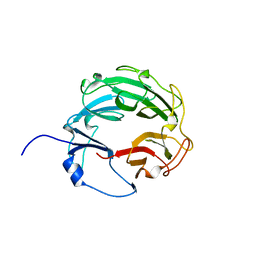 | | Crystal Structure of the Xanthomonas campestris Gyrase A C-terminal Domain | | Descriptor: | DNA gyrase subunit A | | Authors: | Hsieh, T.J, Yen, T.J, Lin, T.S, Chang, H.T, Huang, S.Y, Farh, L, Chan, N.L. | | Deposit date: | 2009-12-26 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Twisting of the DNA binding surface by a beta-strand-bearing proline modulates DNA gyrase activity
To be Published
|
|
4O6G
 
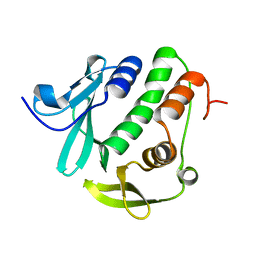 | | Rv3902c from M. tuberculosis | | Descriptor: | Uncharacterized protein | | Authors: | Reddy, B.G, Moates, D.B, Kim, H, Green, T.J, Kim, C, Terwilliger, T.J, Delucas, L.J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-12-20 | | Release date: | 2014-03-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55 angstrom resolution X-ray crystal structure of Rv3902c from Mycobacterium tuberculosis.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4UV4
 
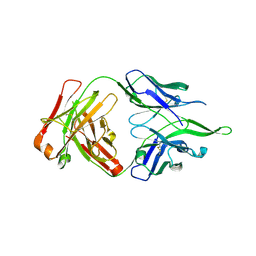 | | Crystal structure of anti-FPR Fpro0165 Fab fragment | | Descriptor: | FPRO0165 FAB | | Authors: | Douthwaite, J.A, Sridharan, S, Huntington, C, Marwood, R, Hammersley, J, Hakulinen, J.K, Ek, M, Sjogren, T, Rider, D, Privezentzev, C, Seaman, J.C, Cariuk, P, Knights, V, Young, J, Wilkinson, T, Sleeman, M, Finch, D.K, Lowe, D.C, Vaughan, T.J. | | Deposit date: | 2014-08-04 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Affinity Maturation of a Novel Antagonistic Human Monoclonal Antibody with a Long Vh Cdr3 Targeting the Class a Gpcr Formyl-Peptide Receptor 1.
Mabs, 7, 2015
|
|
8IQ5
 
 | | Crystal structure of trimeric K2-2 TSP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BICINE, GLYCEROL, ... | | Authors: | Ye, T.J, Huang, K.F, Ko, T.P. | | Deposit date: | 2023-03-15 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Klebsiella pneumoniae K2 capsular polysaccharide degradation by a bacteriophage depolymerase does not require trimer formation.
Mbio, 15, 2024
|
|
7WUK
 
 | | Crystal structure of UBR bof from PRT6 | | Descriptor: | E3 ubiquitin-protein ligase, ZINC ION | | Authors: | Ho, M.C, Lin, T.J. | | Deposit date: | 2022-02-08 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of UBR box from PRT6
To Be Published
|
|
6U1S
 
 | | Cryo-EM structure of a de novo designed 16-helix transmembrane nanopore, TMHC8_R. | | Descriptor: | de novo designed 16-helix transmembrane nanopore, TMHC8_R | | Authors: | Johnson, M.J, Reggiano, G, Xu, C, Lu, P, Hsia, Y, Brunette, T.J, DiMaio, F, Baker, D, Kollman, J. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Computational Design of Transmembrane Channels
To Be Published
|
|
7ABU
 
 | | Structure of SARS-CoV-2 Main Protease bound to RS102895 | | Descriptor: | 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-08 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
5X0I
 
 | |
7A2D
 
 | | Structure-function analyses of dual-BON domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation to the cell division site | | Descriptor: | Uncharacterized protein YraP | | Authors: | Bryant, J.A, Morris, F.C, Knowles, T.J, Maderbocus, R, Heinz, E, Boelter, G, Alodaini, D, Colyer, A, Wotherspoon, P.J, Staunton, K.A, Jeeves, M, Browning, D.F, Sevastsyanovich, Y.R, Wells, T.J, Rossiter, A.E, Bavro, V.N, Sridhar, P, Ward, D.G, Chong, Z.S, Goodall, E.C.A, Icke, C, Teo, A, Chng, S.S, Roper, D.I, Lithgow, T, Cunningham, A.F, Banzhaf, M, Overduin, M, Henderson, I.R. | | Deposit date: | 2020-08-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of dual BON-domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation.
Elife, 9, 2020
|
|
5IWI
 
 | | 1.98A structure of GSK945237 with S.aureus DNA gyrase and singly nicked DNA | | Descriptor: | (1R)-1-[(4-{[(6,7-dihydro[1,4]dioxino[2,3-c]pyridazin-3-yl)methyl]amino}piperidin-1-yl)methyl]-9-fluoro-1,2-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-4-one, CHLORIDE ION, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*AP*CP*CP*GP*CP*AP*CP*A)-3'), ... | | Authors: | Bax, B.D, Miles, T.J. | | Deposit date: | 2016-03-22 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel tricyclics (e.g., GSK945237) as potent inhibitors of bacterial type IIA topoisomerases.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5IWM
 
 | | 2.5A structure of GSK945237 with S.aureus DNA gyrase and DNA. | | Descriptor: | (1R)-1-[(4-{[(6,7-dihydro[1,4]dioxino[2,3-c]pyridazin-3-yl)methyl]amino}piperidin-1-yl)methyl]-9-fluoro-1,2-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-4-one, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*TP*CP*AP*CP*CP*GP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*CP*GP*GP*TP*GP*AP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Bax, B.D, Miles, T.J. | | Deposit date: | 2016-03-22 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel tricyclics (e.g., GSK945237) as potent inhibitors of bacterial type IIA topoisomerases.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
4N34
 
 | | Structure of langerin CRD I313 with alpha-MeGlcNAc | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, methyl 2-acetamido-2-deoxy-alpha-D-glucopyranoside | | Authors: | Feinberg, H, Rowntree, T.J.W, Tan, S.L.W, Drickamer, K, Weis, W.I, Taylor, M.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Common polymorphisms in human langerin change specificity for glycan ligands.
J.Biol.Chem., 288, 2013
|
|
4N8X
 
 | | The structure of Nostoc sp. PCC 7120 CcmL | | Descriptor: | Carbon dioxide concentrating mechanism protein, SULFATE ION | | Authors: | Keeling, T.J, Kimber, M.S. | | Deposit date: | 2013-10-18 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Interactions and structural variability of beta-carboxysomal shell protein CcmL.
Photosynth.Res., 121, 2014
|
|
3PCT
 
 | |
1RVF
 
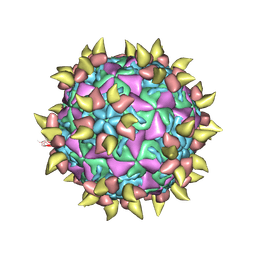 | | FAB COMPLEXED WITH INTACT HUMAN RHINOVIRUS | | Descriptor: | FAB 17-IA, HUMAN RHINOVIRUS 14 COAT PROTEIN | | Authors: | Smith, T.J. | | Deposit date: | 1996-09-05 | | Release date: | 1998-02-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Neutralizing antibody to human rhinovirus 14 penetrates the receptor-binding canyon.
Nature, 383, 1996
|
|
7ADW
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2,4'-Dimethylpropiophenone. | | Descriptor: | 2-methyl-1-(4-methylphenyl)propan-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
4W8J
 
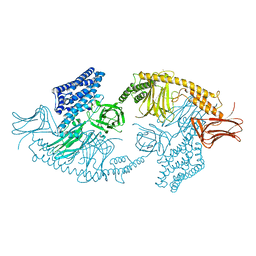 | | Structure of the full-length insecticidal protein Cry1Ac reveals intriguing details of toxin packaging into in vivo formed crystals | | Descriptor: | CALCIUM ION, POTASSIUM ION, Pesticidal crystal protein cry1Ac | | Authors: | Evdokimov, A.G, Moshiri, F, Sturman, E.J, Rydel, T.J, Zheng, M, Seale, J.W, Franklin, S. | | Deposit date: | 2014-08-25 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure of the full-length insecticidal protein Cry1Ac reveals intriguing details of toxin packaging into in vivo formed crystals.
Protein Sci., 23, 2014
|
|
7A1U
 
 | | Structure of SARS-CoV-2 Main Protease bound to Fusidic Acid. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, FUSIDIC ACID, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
4W99
 
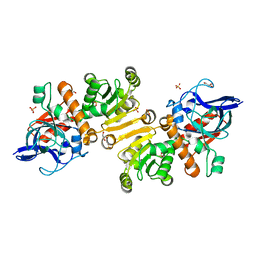 | | Apo-structure of the Y79F,W322E-double mutant of Etr1p | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH, B-specific] 1, mitochondrial, ... | | Authors: | Quade, N, Voegeli, B, Rosenthal, R, Erb, T.J. | | Deposit date: | 2014-08-27 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The use of ene adducts to study and engineer enoyl-thioester reductases.
Nat.Chem.Biol., 11, 2015
|
|
2PXZ
 
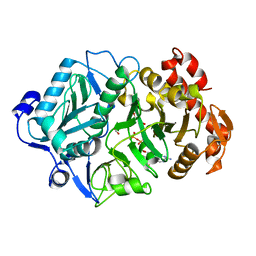 | | E. coli phosphoenolpyruvate carboxykinase (PEPCK) complexed with ATP, Mg2+, Mn2+, carbon dioxide and oxaloacetate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARBON DIOXIDE, MAGNESIUM ION, ... | | Authors: | Delbaere, L.T.J, Cotelesage, J.J.H, Goldie, H. | | Deposit date: | 2007-05-14 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structure of E. coli phosphoenolpyruvate carboxykinase (PEPCK) complexed with ATP, Mg2+, Mn2+, carbon dioxide and oxaloacetate
To be Published
|
|
4UUU
 
 | | 1.7 A resolution structure of human cystathionine beta-synthase regulatory domain (del 516-525) in complex with SAM | | Descriptor: | 1,2-ETHANEDIOL, CYSTATHIONINE BETA-SYNTHASE, S-ADENOSYLMETHIONINE | | Authors: | Kopec, J, McCorvie, T.J, Fitzpatrick, F, Strain-Damerell, C, Froese, D.S, Tallant, C, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2014-07-31 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Inter-Domain Communication of Human Cystathionine Beta Synthase: Structural Basis of S-Adenosyl-L-Methionine Activation.
J.Biol.Chem., 289, 2014
|
|
2PT1
 
 | | FutA1 Synechocystis PCC 6803 | | Descriptor: | Iron transport protein, SULFATE ION | | Authors: | Koropatkin, N.M, Randich, A.M, Bhattachryya-Pakrasi, M, Pakrasi, H.B, Smith, T.J. | | Deposit date: | 2007-05-07 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the iron-binding protein, FutA1, from Synechocystis 6803.
J.Biol.Chem., 282, 2007
|
|
6EQO
 
 | | Tri-functional propionyl-CoA synthase of Erythrobacter sp. NAP1 with bound NADP+ and phosphomethylphosphonic acid adenylate ester | | Descriptor: | Acetyl-coenzyme A synthetase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Zarzycki, J, Bernhardsgruetter, I, Voegeli, B, Wagner, T, Engilberge, S, Girard, E, Shima, S, Erb, T.J. | | Deposit date: | 2017-10-13 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The multicatalytic compartment of propionyl-CoA synthase sequesters a toxic metabolite.
Nat. Chem. Biol., 14, 2018
|
|
6EP8
 
 | | InhA Y158F mutant in complex with NADH from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Enoyl-[acyl-carrier-protein] reductase [NADH], GLYCEROL, ... | | Authors: | Wagner, T, Voegeli, B, Rosenthal, R.G, Stoffel, G, Shima, S, Kiefer, P, Cortina, N, Erb, T.J. | | Deposit date: | 2017-10-11 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | InhA, the enoyl-thioester reductase fromMycobacterium tuberculosisforms a covalent adduct during catalysis.
J. Biol. Chem., 293, 2018
|
|
