8POO
 
 | |
8FYO
 
 | | MicroED structure of Proteinase K from lamellae milled from multiple plasma sources | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Martynowycz, M.W, Shiriaeva, A, Clabbers, M.T.B, Nicolas, W.J, Weaver, S.J, Hattne, J, Gonen, T. | | Deposit date: | 2023-01-26 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.39 Å) | | Cite: | A robust approach for MicroED sample preparation of lipidic cubic phase embedded membrane protein crystals.
Nat Commun, 14, 2023
|
|
7P3S
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 12 | | Descriptor: | GLYCEROL, Histone deacetylase, POTASSIUM ION, ... | | Authors: | Shaik, T.B, Romier, C. | | Deposit date: | 2021-07-08 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.546 Å) | | Cite: | Synthesis, structure-activity relationships, cocrystallization and cellular characterization of novel smHDAC8 inhibitors for the treatment of schistosomiasis.
Eur.J.Med.Chem., 225, 2021
|
|
6MAC
 
 | | Ternary structure of GDF11 bound to ActRIIB-ECD and Alk5-ECD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2B, Growth/differentiation factor 11, ... | | Authors: | Goebel, E.J, Thompson, T.B. | | Deposit date: | 2018-08-27 | | Release date: | 2019-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural characterization of an activin class ternary receptor complex reveals a third paradigm for receptor specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6EVI
 
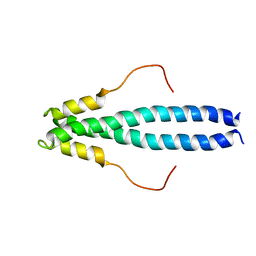 | | solution NMR structure of EB1 C terminus (191-260) | | Descriptor: | Microtubule-associated protein RP/EB family member 1 | | Authors: | Barsukov, I.L, Almeida, T.B. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Targeting SxIP-EB1 interaction: An integrated approach to the discovery of small molecule modulators of dynamic binding sites.
Sci Rep, 7, 2017
|
|
5GPO
 
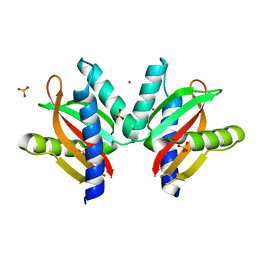 | | The sensor domain structure of the zinc-responsive histidine kinase CzcS from Pseudomonas Aeruginosa | | Descriptor: | SULFATE ION, Sensor protein CzcS, ZINC ION | | Authors: | Wang, D, Chen, W.Z, Huang, S.Q, Liu, X.C, Hu, Q.Y, Wei, T.B, Gan, J.H, Chen, H. | | Deposit date: | 2016-08-03 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structural basis of Zn(II) induced metal detoxification and antibiotic resistance by histidine kinase CzcS in Pseudomonas aeruginosa
PLoS Pathog., 13, 2017
|
|
9FGE
 
 | |
5BN6
 
 | | Crystal Structure of Frutalin from Artocarpus incisa in complex with galactose | | Descriptor: | Jacalin, beta-D-galactopyranose | | Authors: | Vieira Neto, A.E, Pereira, H.M, Moreno, F.B.M.B, Moreira, A.C.O.M, Lobo, M.D.P, Sousa, F.D, Grangeiro, T.B, Moreira, R.A. | | Deposit date: | 2015-05-25 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6499 Å) | | Cite: | Crystal Structure of Frutalin from Artocarpus incisa in complex with galactose
To Be Published
|
|
5ZXB
 
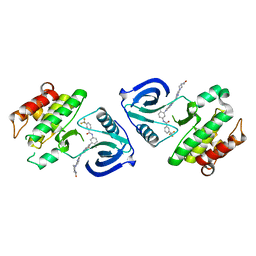 | | Crystal structure of ACK1 with compound 10d | | Descriptor: | Activated CDC42 kinase 1, N-{3-[7-{[6-(4-acetylpiperazin-1-yl)pyridin-3-yl]amino}-1-methyl-2-oxo-1,4-dihydropyrimido[4,5-d]pyrimidin-3(2H)-yl]-4-methylphenyl}-3-(trifluoromethyl)benzamide | | Authors: | Hong, E.M, Kim, H.L, Sim, T.B. | | Deposit date: | 2018-05-18 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | First SAR Study for Overriding NRAS Mutant Driven Acute Myeloid Leukemia.
J. Med. Chem., 61, 2018
|
|
7ULY
 
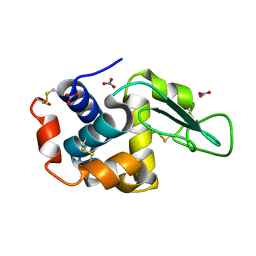 | | MicroED structure of triclinic lysozyme | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Clabbers, M.T.B, Martynowycz, M.W, Hattne, J, Gonen, T. | | Deposit date: | 2022-04-05 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-09 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.87 Å) | | Cite: | Hydrogens and hydrogen-bond networks in macromolecular MicroED data.
J Struct Biol X, 6, 2022
|
|
6MAA
 
 | | WFIKKN2 Follistatin Domain | | Descriptor: | NITRATE ION, WAP, Kazal, ... | | Authors: | McCoy, J.C, Walker, R.G, Thomas, T.B. | | Deposit date: | 2018-08-27 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.394 Å) | | Cite: | Crystal structure of the WFIKKN2 follistatin domain reveals insight into how it inhibits growth differentiation factor 8 (GDF8) and GDF11.
J.Biol.Chem., 294, 2019
|
|
3UF1
 
 | | Crystal Structure of Vimentin (fragment 144-251) from Homo sapiens, Northeast Structural Genomics Consortium Target HR4796B | | Descriptor: | Vimentin | | Authors: | Kuzin, A, Abashidze, M, Vorobiev, S.M, Patel, P, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-10-31 | | Release date: | 2011-11-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The structure of vimentin linker 1 and rod 1B domains characterized by site-directed spin-labeling electron paramagnetic resonance (SDSL-EPR) and X-ray crystallography.
J.Biol.Chem., 287, 2012
|
|
4NVS
 
 | | Crystal Structure of the Q18CP6_CLOD6 protein from glyoxalase family. Northeast Structural Genomics Consortium Target CfR3 | | Descriptor: | Putative enzyme, glyoxalase family | | Authors: | Vorobiev, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-12-05 | | Release date: | 2013-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | Crystal Structure of the Q18CP6_CLOD6 protein from glyoxalase family.
To be Published
|
|
4WOG
 
 | | Crystal Structure of Frutalin from Artocarpus incisa | | Descriptor: | Frutalin | | Authors: | Pereira, H.M, Moreira, A.C.O.M, Vieira Neto, A.E, Moreno, F.B.M.B, Lobo, M.D.P, Sousa, F.D, Grangeiro, T.B, Moreira, R.A. | | Deposit date: | 2014-10-15 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.813 Å) | | Cite: | Crystal Structure of Frutalin from Artocarpus incisa
To Be Published
|
|
6VFM
 
 | | Crystal structure of SpeG allosteric polyamine acetyltransferase from Bacillus thuringiensis | | Descriptor: | Spermidine N1-acetyltransferase | | Authors: | Tsimbalyuk, S, Shornikov, A, Le, V.T.B, Kuhn, M.L, Forwood, J.K. | | Deposit date: | 2020-01-05 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | SpeG polyamine acetyltransferase enzyme from Bacillus thuringiensis forms a dodecameric structure and exhibits high catalytic efficiency.
J.Struct.Biol., 210, 2020
|
|
3USH
 
 | | Crystal Structure of the Q2S0R5 protein from Salinibacter ruber, Northeast Structural Genomics Consortium Target SrR207 | | Descriptor: | BROMIDE ION, Uncharacterized protein | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Crystal Structure of the Q2S0R5 protein from Salinibacter ruber, Northeast Structural Genomics Consortium Target SrR207
To be Published
|
|
4NZG
 
 | | Crystal Structure of the N-terminal domain of Moloney murine leukemia virus integrase, Northeast Structural Genomics Consortium Target OR3 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETATE ION, Integrase p46, ... | | Authors: | Guan, R, Jiang, M, Janjua, H, Maglaqui, M, Zhao, L, Xiao, R, Acton, T.B, Everett, J.K, Roth, M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-12-12 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | X-ray crystal structure of the N-terminal region of Moloney murine leukemia virus integrase and its implications for viral DNA recognition.
Proteins, 85, 2017
|
|
4X1J
 
 | | X-ray crystal structure of the dimeric BMP antagonist NBL1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuroblastoma suppressor of tumorigenicity 1 | | Authors: | Thompson, T.B, Nolan, K, Kattamuri, C. | | Deposit date: | 2014-11-24 | | Release date: | 2015-01-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Neuroblastoma Suppressor of Tumorigenicity 1 (NBL1): INSIGHTS FOR THE FUNCTIONAL VARIABILITY ACROSS BONE MORPHOGENETIC PROTEIN (BMP) ANTAGONISTS.
J.Biol.Chem., 290, 2015
|
|
6VFN
 
 | | Crystal structure of SpeG allosteric polyamine acetyltransferase from Bacillus thuringiensis in complex with spermine | | Descriptor: | SPERMINE, Spermidine N1-acetyltransferase | | Authors: | Tsimbalyuk, S, Shornikov, A, Le, V.T.B, Kuhn, M.L, Forwood, J.K. | | Deposit date: | 2020-01-05 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | SpeG polyamine acetyltransferase enzyme from Bacillus thuringiensis forms a dodecameric structure and exhibits high catalytic efficiency.
J.Struct.Biol., 210, 2020
|
|
8E52
 
 | | MicroED structure of proteinase K recorded on K2 | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Clabbers, M.T.B, Martynowycz, M.W, Hattne, J, Nannenga, B.L, Gonen, T. | | Deposit date: | 2022-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | Cite: | Electron-counting MicroED data with the K2 and K3 direct electron detectors.
J.Struct.Biol., 214, 2022
|
|
8E54
 
 | | MicroED structure of triclinic lysozyme recorded on K3 | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Clabbers, M.T.B, Martynowycz, M.W, Hattne, J, Nannenga, B.L, Gonen, T. | | Deposit date: | 2022-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.2 Å) | | Cite: | Electron-counting MicroED data with the K2 and K3 direct electron detectors.
J.Struct.Biol., 214, 2022
|
|
8E53
 
 | | MicroED structure of proteinase K recorded on K3 | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Clabbers, M.T.B, Martynowycz, M.W, Hattne, J, Nannenga, B.L, Gonen, T. | | Deposit date: | 2022-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.7 Å) | | Cite: | Electron-counting MicroED data with the K2 and K3 direct electron detectors.
J.Struct.Biol., 214, 2022
|
|
5DD1
 
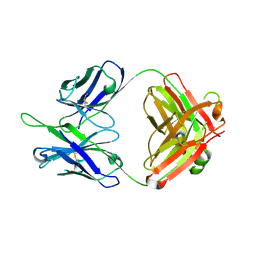 | | Crystal structures in an anti-HIV antibody lineage from immunization of Rhesus macaques | | Descriptor: | ANTI-HIV ANTIBODY DH570 FAB HEAVY CHAIN, ANTI-HIV ANTIBODY DH570 FAB LIGHT CHAIN | | Authors: | Zhang, R, Verkoczy, L, Wiehe, K, Alam, S.M, Nicely, N.I, Santra, S, Bradley, T, Pemble, C, Gao, F, Montefiori, D.C, Bouton-Verville, H, Kelsoe, G, Parks, R, Foulger, A, Tomaras, G, Keple, T.B, Moody, M.A, Liao, H.-X, Haynes, B.F. | | Deposit date: | 2015-08-24 | | Release date: | 2016-05-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Initiation of immune tolerance-controlled HIV gp41 neutralizing B cell lineages.
Sci Transl Med, 8, 2016
|
|
7Z7D
 
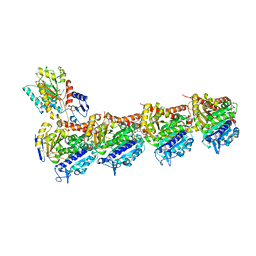 | | Tubulin-Todalam-Vinblastine-complex | | Descriptor: | (2ALPHA,2'BETA,3BETA,4ALPHA,5BETA)-VINCALEUKOBLASTINE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Muehlethaler, T, Milanos, L, Ortega, J.A, Blum, T.B, Gioia, D, Roy, B, Prota, A.E, Cavalli, A, Steinmetz, M.O. | | Deposit date: | 2022-03-15 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational Design of a Novel Tubulin Inhibitor with a Unique Mechanism of Action.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7Z5S
 
 | | Crystal Structure of botulinum neurotoxin A2 cell binding domain in complex with GD1a | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin, HEXAETHYLENE GLYCOL, ... | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M, Mahadeva, T.B. | | Deposit date: | 2022-03-10 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Features of Clostridium botulinum Neurotoxin Subtype A2 Cell Binding Domain.
Toxins, 14, 2022
|
|
