4URT
 
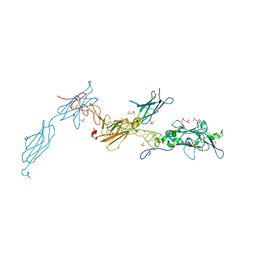 | | The crystal structure of a fragment of netrin-1 in complex with FN5- FN6 of DCC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Finci, L.I, Krueger, N, Sun, X, Zhang, J, Chegkazi, M, Wu, Y, Schenk, G, Mertens, H.D.T, Svergun, D.I, Zhang, Y, Wang, J.-h, Meijers, R. | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Crystal Structure of Netrin-1 in Complex with Dcc Reveals the Bi-Functionality of Netrin-1 as a Guidance Cue
Neuron, 83, 2014
|
|
3ZLJ
 
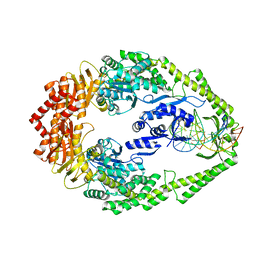 | | CRYSTAL STRUCTURE OF FULL-LENGTH E.COLI DNA MISMATCH REPAIR PROTEIN MUTS D835R MUTANT IN COMPLEX WITH GT MISMATCHED DNA | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP *AP*GP*TP*GP*TP*CP*AP)-3', 5'-D(*TP*GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP *GP*GP*CP*AP*GP*CP*TP)-3', DNA MISMATCH REPAIR PROTEIN MUTS | | Authors: | Groothuizen, F.S, Fish, A, Petoukhov, M.V, Reumer, A, Manelyte, L, Winterwerp, H.H.K, Marinus, M.G, Lebbink, J.H.G, Svergun, D.I, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2013-02-01 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Using Stable Muts Dimers and Tetramers to Quantitatively Analyze DNA Mismatch Recognition and Sliding Clamp Formation.
Nucleic Acids Res., 41, 2013
|
|
4WUO
 
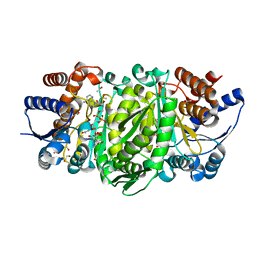 | | Structure of the E270A Mutant Isopropylmalate dehydrogenase from Thermus thermophilus in complex with IPM, Mn and NADH | | Descriptor: | 3-ISOPROPYLMALIC ACID, 3-isopropylmalate dehydrogenase, ETHANOL, ... | | Authors: | Pallo, A, Graczer, E, Olah, J, Szimler, T, Konarev, P.V, Svergun, D.I, Merli, A, Zavodszky, P, Vas, M, Weiss, M.S. | | Deposit date: | 2014-11-03 | | Release date: | 2014-11-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Glutamate 270 plays an essential role in K(+)-activation and domain closure of Thermus thermophilus isopropylmalate dehydrogenase.
Febs Lett., 589, 2015
|
|
4HKQ
 
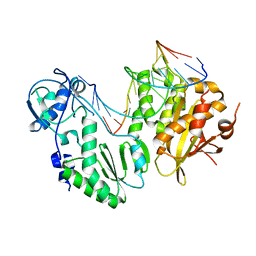 | | XMRV reverse transcriptase in complex with RNA/DNA hybrid | | Descriptor: | DNA (5'-D(*TP*GP*GP*AP*AP*TP*CP*A*GP*GP*TP*GP*TP*CP*GP*CP*AP*CP*TP*CP*TP*G)-3'), RNA (5'-R(*AP*AP*CP*AP*GP*AP*GP*UP*GP*CP*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*CP*CP*AP*U)-3'), Reverse transcriptase/ribonuclease H p80 | | Authors: | Nowak, E, Potrzebowski, W, Konarev, P.V, Rausch, J.W, Bona, M.K, Svergun, D.I, Bujnicki, J.M, Le Grice, S.F.J, Nowotny, M. | | Deposit date: | 2012-10-15 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural analysis of monomeric retroviral reverse transcriptase in complex with an RNA/DNA hybrid
Nucleic Acids Res., 41, 2013
|
|
1EG0
 
 | | FITTING OF COMPONENTS WITH KNOWN STRUCTURE INTO AN 11.5 A CRYO-EM MAP OF THE E.COLI 70S RIBOSOME | | Descriptor: | FORMYL-METHIONYL-TRNA, FRAGMENT OF 16S RRNA HELIX 23, FRAGMENT OF 23S RRNA, ... | | Authors: | Gabashvili, I.S, Agrawal, R.K, Spahn, C.M.T, Grassucci, R.A, Svergun, D.I, Frank, J, Penczek, P. | | Deposit date: | 2000-02-11 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Solution structure of the E. coli 70S ribosome at 11.5 A resolution.
Cell(Cambridge,Mass.), 100, 2000
|
|
4OFS
 
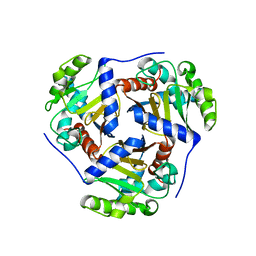 | | Crystal structure of a truncated catalytic core of the 2-oxoacid dehydrogenase multienzyme complex from Thermoplasma acidophilum | | Descriptor: | Probable lipoamide acyltransferase | | Authors: | Marrot, N.L, Marshall, J.J.T, Svergun, D.I, Crennell, S.J, Hough, D.W, van den Elsen, J.M.H, Danson, M.J. | | Deposit date: | 2014-01-15 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Why are the 2-oxoacid dehydrogenase complexes so large? Generation of an active trimeric complex.
Biochem.J., 463, 2014
|
|
2KMS
 
 | | Combined high- and low-resolution techniques reveal compact structure in central portion of factor H despite long inter-modular linkers | | Descriptor: | Complement factor H | | Authors: | Schmidt, C.Q, Herbert, A.P, Guariento, M, Mertens, H.D.T, Soares, D.C, Uhrin, D, Rowe, A.J, Svergun, D.I, Barlow, P.N. | | Deposit date: | 2009-08-04 | | Release date: | 2009-11-03 | | Last modified: | 2020-02-26 | | Method: | SOLUTION NMR | | Cite: | The Central Portion of Factor H (Modules 10-15) Is Compact and Contains a Structurally Deviant CCP Module
J.Mol.Biol., 395, 2010
|
|
6ZJ4
 
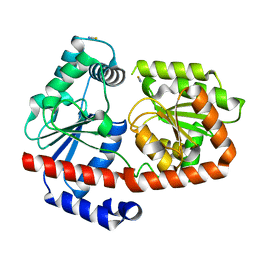 | | apo-Trehalose transferase (apo-TreT) from Thermoproteus uzoniensis | | Descriptor: | THIOCYANATE ION, Trehalose phosphorylase/synthase | | Authors: | Bento, I, Mestrom, L, Marsden, S.R, van der Eijk, H, Laustsen, J.U, Jeffries, C.M, Svergun, D.I, Hagedoorn, P.-H, Hanefeld, U. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Anomeric Selectivity of Trehalose Transferase with Rare l-Sugars.
Acs Catalysis, 10, 2020
|
|
6ZJH
 
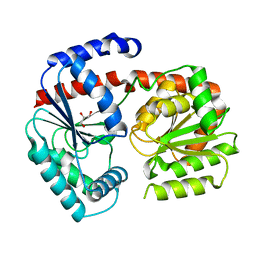 | | Trehalose transferase from Thermoproteus uzoniensis soaked with trehalose | | Descriptor: | GLYCEROL, Trehalose phosphorylase/synthase, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Bento, I, Mestrom, L, Marsden, S.R, van der Eijk, H, Laustsen, J.U, Jeffries, C.M, Svergun, D.I, Hagedoorn, P.-H, Hanefeld, U. | | Deposit date: | 2020-06-29 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Anomeric Selectivity of Trehalose Transferase with Rare l-Sugars.
Acs Catalysis, 10, 2020
|
|
6ZJ7
 
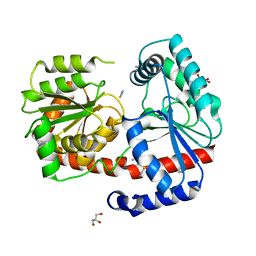 | | Trehalose transferase (TreT) from Thermoproteus uzoniensis soaked with Mg | | Descriptor: | GLYCEROL, TETRAETHYLENE GLYCOL, THIOCYANATE ION, ... | | Authors: | Bento, I, Mestrom, L, Marsden, S.R, van der Eijk, H, Laustsen, J.U, Jeffries, C.M, Svergun, D.I, Hagedoorn, P.-H, Hanefeld, U. | | Deposit date: | 2020-06-28 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Anomeric Selectivity of Trehalose Transferase with Rare l-Sugars.
Acs Catalysis, 10, 2020
|
|
6ZMZ
 
 | | UDPG-bound Trehalose transferase from Thermoproteus uzoniensis | | Descriptor: | DI(HYDROXYETHYL)ETHER, Trehalose phosphorylase/synthase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Bento, I, Mestrom, L, Marsden, S.R, van der Eijk, H, Laustsen, J.U, Jeffries, C.M, Svergun, D.I, Hagedoorn, P.-H, Hanefeld, U. | | Deposit date: | 2020-07-05 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Anomeric Selectivity of Trehalose Transferase with Rare l-Sugars.
Acs Catalysis, 10, 2020
|
|
6ZN1
 
 | | Trehalose transferase bound to alpha-D-glucopyranosyl-beta-galactopyranose from Thermoproteus uzoniensis | | Descriptor: | DI(HYDROXYETHYL)ETHER, THIOCYANATE ION, Trehalose phosphorylase/synthase, ... | | Authors: | Bento, I, Mestrom, L, Marsden, S.R, van der Eijk, H, Laustsen, J.U, Jeffries, C.M, Svergun, D.I, Hagedoorn, P.-H, Hanefeld, U. | | Deposit date: | 2020-07-06 | | Release date: | 2021-01-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Anomeric Selectivity of Trehalose Transferase with Rare l-Sugars.
ACS Catal, 10, 2020
|
|
2JQR
 
 | | Solution model of crosslinked complex of cytochrome c and adrenodoxin | | Descriptor: | Adrenodoxin, mitochondrial, Cytochrome c iso-1, ... | | Authors: | Xu, X, Reinle, W, Hannemann, F, Konarev, P.V, Svergun, D.I, Bernhardt, R, Ubbink, M. | | Deposit date: | 2007-06-07 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Dynamics in a pure encounter complex of two proteins studied by solution scattering and paramagnetic NMR spectroscopy
J.Am.Chem.Soc., 130, 2008
|
|
3HS0
 
 | | Cobra Venom Factor (CVF) in complex with human factor B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cobra venom factor, ... | | Authors: | Janssen, B.J.C, Gomes, L, Koning, R.I, Svergun, D.I, Koster, A.J, Fritzinger, D.C, Vogel, C.-W, Gros, P. | | Deposit date: | 2009-06-10 | | Release date: | 2009-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights into complement convertase formation based on the structure of the factor B-cobra venom factor complex
Embo J., 28, 2009
|
|
3HRZ
 
 | | Cobra Venom Factor (CVF) in complex with human factor B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cobra venom factor, ... | | Authors: | Janssen, B.J.C, Gomes, L, Koning, R.I, Svergun, D.I, Koster, A.J, Fritzinger, D.C, Vogel, C.-W, Gros, P. | | Deposit date: | 2009-06-10 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into complement convertase formation based on the structure of the factor B-cobra venom factor complex
Embo J., 28, 2009
|
|
5A6S
 
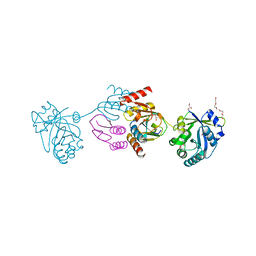 | | Crystal structure of the CTP1L endolysin reveals how its activity is regulated by a secondary translation product | | Descriptor: | ENDOLYSIN, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Dunne, M, Leicht, S, Krichel, B, Mertens, H.D.T, Thompson, A, Krijgsveld, J, Svergun, D.I, GomezTorres, N, Garde, S, Uetrecht, C, Narbad, A, Mayer, M.J, Meijers, R. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Ctp1L Endolysin Reveals How its Activity is Regulated by a Secondary Translation Product.
J.Biol.Chem., 291, 2016
|
|
5AD2
 
 | | Bivalent binding to BET bromodomains | | Descriptor: | (3R)-4-(2-{4-[1-(3-chloro[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidinyl]phenoxy}ethyl)-1,3-dimethyl-2-piperazinone, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Waring, M.J, Chen, H, Rabow, A.A, Walker, G, Bobby, R, Boiko, S, Bradbury, R.H, Callis, R, Dale, I, Daniels, D, Flavell, L, Holdgate, G, Jowitt, T.A, Kikhney, A, McAlister, M, Ogg, D, Patel, J, Petteruti, P, Robb, G.R, Robers, M, Stratton, N, Svergun, D.I, Wang, W, Whittaker, D. | | Deposit date: | 2015-08-19 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Potent and Selective Bivalent Inhibitors of Bet Bromodomains
Nat.Chem.Biol., 12, 2016
|
|
5AD3
 
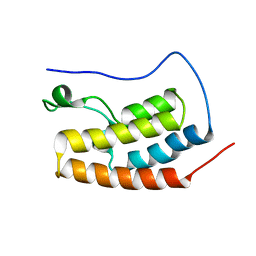 | | Bivalent binding to BET bromodomains | | Descriptor: | 3-methoxy-N-[2-[4-[1-(3-methoxy-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidyl]phenoxy]ethyl]-N-methyl-[1,2,4]triazolo[4,3-b]pyridazin-6-amine, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Waring, M.J, Chen, H, Rabow, A.A, Walker, G, Bobby, R, Boiko, S, Bradbury, R.H, Callis, R, Dale, I, Daniels, D, Flavell, L, Holdgate, G, Jowitt, T.A, Kikhney, A, McAlister, M, Ogg, D, Patel, J, Petteruti, P, Robb, G.R, Robers, M, Stratton, N, Svergun, D.I, Wang, W, Whittaker, D. | | Deposit date: | 2015-08-19 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Potent and Selective Bivalent Inhibitors of Bet Bromodomains
Nat.Chem.Biol., 12, 2016
|
|
2VDC
 
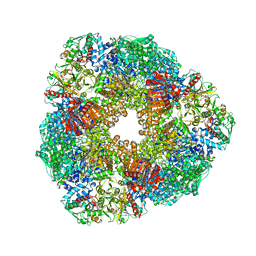 | | THE 9.5 A RESOLUTION STRUCTURE OF GLUTAMATE SYNTHASE FROM CRYO-ELECTRON MICROSCOPY AND ITS OLIGOMERIZATION BEHAVIOR IN SOLUTION: FUNCTIONAL IMPLICATIONS. | | Descriptor: | 2-OXOGLUTARIC ACID, FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Cottevieille, M, Larquet, E, Jonic, S, Petoukhov, M.V, Caprini, G, Paravisi, S, Svergun, D.I, Vanoni, M.A, Boisset, N. | | Deposit date: | 2007-10-04 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | The Subnanometer Resolution Structure of the Glutamate Synthase 1.2-Mda Hexamer by Cryoelectron Microscopy and its Oligomerization Behavior in Solution: Functional Implications.
J.Biol.Chem., 283, 2008
|
|
2XWX
 
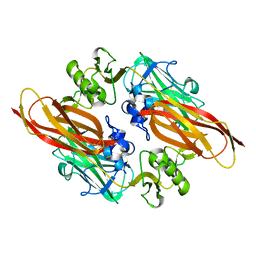 | | Vibrio cholerae colonization factor GbpA crystal structure | | Descriptor: | GLCNAC-BINDING PROTEIN A | | Authors: | Wong, E, Vaaje-Kolstad, G, Ghosh, A, Guerrero, R.H, Konarev, P.V, Ibrahim, A.F.M, Svergun, D.I, Eijsink, V.G.H, Chatterjee, N.S, van Aalten, D.M.F. | | Deposit date: | 2010-11-06 | | Release date: | 2011-11-16 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Vibrio Cholerae Colonization Factor Gbpa Possesses a Modular Structure that Governs Binding to Different Host Surfaces.
Plos Pathog., 8, 2012
|
|
2XSZ
 
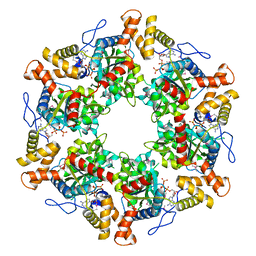 | | The dodecameric human RuvBL1:RuvBL2 complex with truncated domains II | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, RUVB-LIKE 1, RUVB-LIKE 2 | | Authors: | Gorynia, S, Bandeiras, T.M, Matias, P.M, Pinho, F.G, McVey, C.E, Vonrhein, C, Svergun, D.I, Round, A, Donner, P, Carrondo, M.A. | | Deposit date: | 2010-10-01 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Insights Into a Dodecameric Molecular Machine - the Ruvbl1/Ruvbl2 Complex.
J.Struct.Biol., 176, 2011
|
|
3BA0
 
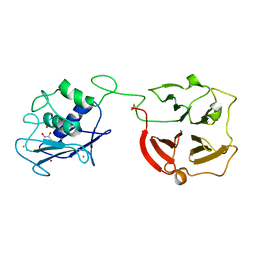 | | Crystal structure of full-length human MMP-12 | | Descriptor: | ACETOHYDROXAMIC ACID, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Bertini, I, Calderone, V, Fragai, M, Jaiswal, R, Luchinat, C, Melikian, M, Myonas, E, Svergun, D.I. | | Deposit date: | 2007-11-07 | | Release date: | 2008-07-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Evidence of reciprocal reorientation of the catalytic and hemopexin-like domains of full-length MMP-12.
J.Am.Chem.Soc., 130, 2008
|
|
1SD5
 
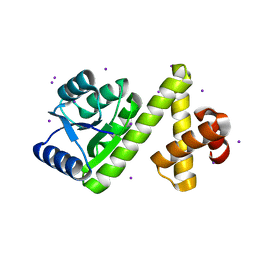 | | Crystal structure of Rv1626 | | Descriptor: | IODIDE ION, putative antiterminator | | Authors: | Morth, J.P, Feng, V, Perry, L.J, Svergun, D.I, Tucker, P.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-02-13 | | Release date: | 2004-09-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Crystal and Solution Structure of a Putative Transcriptional Antiterminator from Mycobacterium tuberculosis.
Structure, 12, 2004
|
|
1SJQ
 
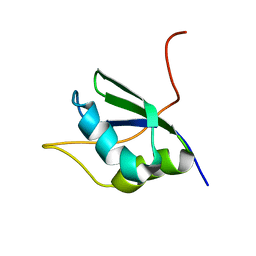 | | NMR Structure of RRM1 from Human Polypyrimidine Tract Binding Protein Isoform 1 (PTB1) | | Descriptor: | Polypyrimidine tract-binding protein 1 | | Authors: | Simpson, P.J, Monie, T.P, Szendroi, A, Davydova, N, Tyzack, J.K, Conte, M.R, Read, C.M, Cary, P.D, Svergun, D.I, Konarev, P.V, Petoukhov, M.V, Curry, S, Matthews, S.J. | | Deposit date: | 2004-03-04 | | Release date: | 2004-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and RNA Interactions of the N-Terminal RRM Domains of PTB
Structure, 12, 2004
|
|
1S8N
 
 | | Crystal structure of Rv1626 from Mycobacterium tuberculosis | | Descriptor: | AZIDE ION, putative antiterminator | | Authors: | Morth, J.P, Feng, V, Perry, L.J, Svergun, D.I, Tucker, P.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-02-03 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | The Crystal and Solution Structure of a Putative Transcriptional Antiterminator from Mycobacterium tuberculosis.
Structure, 12, 2004
|
|
