5WRA
 
 | | Crystal structure of hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Sugahara, M, Suzuki, M, Masuda, T, Inoue, S, Nango, E. | | Deposit date: | 2016-12-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Hydroxyethyl cellulose matrix applied to serial crystallography
Sci Rep, 7, 2017
|
|
5WRB
 
 | | Crystal structure of hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Sugahara, M, Suzuki, M, Masuda, T, Inoue, S, Nango, E. | | Deposit date: | 2016-12-01 | | Release date: | 2017-12-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.013 Å) | | Cite: | Hydroxyethyl cellulose matrix applied to serial crystallography
Sci Rep, 7, 2017
|
|
1O3Y
 
 | | Crystal structure of mouse ARF1 (delta17-Q71L), GTP form | | Descriptor: | ADP-ribosylation factor 1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Shiba, T, Kawasaki, M, Takatsu, H, Nogi, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2003-05-08 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport
Nat.Struct.Biol., 10, 2003
|
|
5WRC
 
 | | Crystal structure of proteinase K from Engyodontium album | | Descriptor: | NITRATE ION, PRASEODYMIUM ION, Proteinase K | | Authors: | Sugahara, M, Nakane, T, Suzuki, M, Masuda, T, Inoue, S, Numata, K. | | Deposit date: | 2016-12-01 | | Release date: | 2017-11-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hydroxyethyl cellulose matrix applied to serial crystallography
Sci Rep, 7, 2017
|
|
2GCC
 
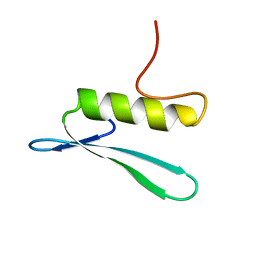 | | SOLUTION STRUCTURE OF THE GCC-BOX BINDING DOMAIN, NMR, MINIMIZED MEAN STRUCTURE | | Descriptor: | ATERF1 | | Authors: | Allen, M.D, Yamasaki, K, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | Deposit date: | 1998-03-13 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
5Y5N
 
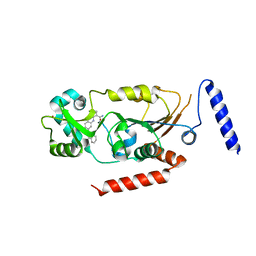 | | Crystal structure of human Sirtuin 2 in complex with a selective inhibitor | | Descriptor: | 2-[[3-(2-phenylethoxy)phenyl]amino]benzamide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION | | Authors: | Mellini, P, Itoh, Y, Tsumoto, H, Li, Y, Suzuki, M, Tokuda, N, Kakizawa, T, Miura, Y, Takeuchi, J, Lahtela-Kakkonen, M, Suzuki, T. | | Deposit date: | 2017-08-09 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potent mechanism-based sirtuin-2-selective inhibition by anin situ-generated occupant of the substrate-binding site, "selectivity pocket" and NAD+-binding site.
Chem Sci, 8, 2017
|
|
1HI3
 
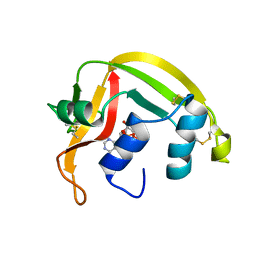 | | Eosinophil-derived Neurotoxin (EDN) - Adenosine 2'-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
1HI4
 
 | | Eosinophil-derived Neurotoxin (EDN) - Adenosien-3'-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
1HI2
 
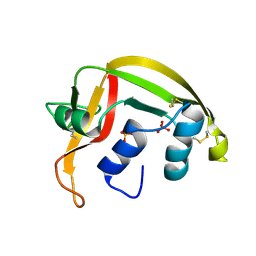 | | Eosinophil-derived Neurotoxin (EDN) - Sulphate Complex | | Descriptor: | EOSINOPHIL-DERIVED NEUROTOXIN, SULFATE ION | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
1HI5
 
 | | Eosinophil-derived Neurotoxin (EDN) - Adenosine-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
5SW0
 
 | |
5SW2
 
 | | Thaumatin Structure at pH 6.0, orthorhombic type1 | | Descriptor: | GLYCEROL, Thaumatin I | | Authors: | Masuda, T, Sano, A, Murata, K, Okubo, K, Suzuki, M, Mikami, B. | | Deposit date: | 2016-08-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Thaumatin Structure at pH 6.0, orthorhombic type1
To Be Published
|
|
5SW1
 
 | | Thaumatin Structure at pH 6.0 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Thaumatin I | | Authors: | Masuda, T, Sano, A, Murata, K, Okubo, K, Suzuki, M, Mikami, B. | | Deposit date: | 2016-08-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Thaumatin Structure at pH 6.0
To Be Published
|
|
5D4H
 
 | | High-resolution nitrite complex of a copper nitrite reductase determined by synchrotron radiation crystallography | | Descriptor: | ACETIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5D4I
 
 | | Intact nitrite complex of a copper nitrite reductase determined by serial femtosecond crystallography | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5D4J
 
 | | Chloride-bound form of a copper nitrite reductase from Alcaligenes faecals | | Descriptor: | ACETIC ACID, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5F7A
 
 | | Nitrite complex structure of copper nitrite reductase from Alcaligenes faecalis determined at 293 K | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-12-07 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5F7B
 
 | | Resting state structure of CuNiR form Alcaligenes faecalis determined at 293 K | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-12-07 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1RI7
 
 | |
7CE4
 
 | | Tankyrase2 catalytic domain in complex with K-476 | | Descriptor: | 5-[3-[[1-(6,7-dimethoxyquinazolin-4-yl)piperidin-4-yl]methyl]-2-oxidanylidene-4H-quinazolin-1-yl]-2-fluoranyl-benzenecarbonitrile, Poly [ADP-ribose] polymerase tankyrase-2, SULFATE ION, ... | | Authors: | Takahashi, Y, Suzuki, M, Saito, J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The dual pocket binding novel tankyrase inhibitor K-476 enhances the efficacy of immune checkpoint inhibitor by attracting CD8 + T cells to tumors.
Am J Cancer Res, 11, 2021
|
|
2K7W
 
 | | BAX Activation is Initiated at a Novel Interaction Site | | Descriptor: | Apoptosis regulator BAX, Bcl-2-like protein 11 | | Authors: | Gavathiotis, E, Suzuki, M, Davis, M.L, Pitter, K, Bird, G.H, Katz, S.G, Tu, H.C, Kim, H, Cheng, E.H, Tjandra, N, Walensky, L.D. | | Deposit date: | 2008-08-27 | | Release date: | 2008-10-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | BAX activation is initiated at a novel interaction site.
Nature, 455, 2008
|
|
2HI7
 
 | | Crystal structure of DsbA-DsbB-ubiquinone complex | | Descriptor: | Disulfide bond formation protein B, Thiol:disulfide interchange protein dsbA, UBIQUINONE-1, ... | | Authors: | Inaba, K, Murakami, S, Suzuki, M, Nakagawa, A, Yamashita, E, Okada, K, Ito, K. | | Deposit date: | 2006-06-29 | | Release date: | 2006-12-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of the DsbB-DsbA Complex Reveals a Mechanism of Disulfide Bond Generation
Cell(Cambridge,Mass.), 127, 2006
|
|
5Y1A
 
 | | HBP35 of Porphyromonas gingivalis | | Descriptor: | 35 kDa hemin binding protein | | Authors: | Kakuda, S, Suzuki, M, Sato, K. | | Deposit date: | 2017-07-20 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Immunoglobulin-like domains of the cargo proteins are essential for protein stability during secretion by the type IX secretion system.
Mol. Microbiol., 110, 2018
|
|
1JWG
 
 | | VHS Domain of human GGA1 complexed with cation-independent M6PR C-terminal Peptide | | Descriptor: | ADP-ribosylation factor binding protein GGA1, Cation-independent mannose-6-phosphate receptor, IODIDE ION | | Authors: | Shiba, T, Takatsu, H, Nogi, T, Matsugaki, N, Kawasaki, M, Igarashi, N, Suzuki, M, Kato, R, Earnest, T, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2001-09-04 | | Release date: | 2002-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for recognition of acidic-cluster dileucine sequence by GGA1.
Nature, 415, 2002
|
|
1JWF
 
 | | Crystal Structure of human GGA1 VHS domain. | | Descriptor: | ADP-ribosylation factor binding protein GGA1 | | Authors: | Shiba, T, Takatsu, H, Nogi, T, Matsugaki, N, Kawasaki, M, Igarashi, N, Suzuki, M, Kato, R, Earnest, T, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2001-09-04 | | Release date: | 2002-03-06 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for recognition of acidic-cluster dileucine sequence by GGA1.
Nature, 415, 2002
|
|
