3PSP
 
 | |
8WQW
 
 | | Cryo-EM structure of bsAb3 Fab-Gn-Gc complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein C, Glycoprotein N, ... | | Authors: | Wu, Y, Sun, J.Q. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Bispecific antibodies targeting two glycoproteins on SFTSV exhibit synergistic neutralization and protection in a mouse model.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8XS3
 
 | | Structure of MPXV B6 and D68 fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D68_heavy chain, ... | | Authors: | wu, L.L, Sun, J.Q. | | Deposit date: | 2024-01-08 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Two noncompeting human neutralizing antibodies targeting MPXV B6 show protective effects against orthopoxvirus infections.
Nat Commun, 15, 2024
|
|
3UFB
 
 | | Crystal structure of a modification subunit of a putative type I restriction enzyme from Vibrio vulnificus YJ016 | | Descriptor: | Type I restriction-modification system methyltransferase subunit | | Authors: | Park, S.Y, Lee, H.J, Sun, J, Nishi, K, Song, J.M, Kim, J.S. | | Deposit date: | 2011-11-01 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of a modification subunit of a putative type I restriction enzyme from Vibrio vulnificus YJ016
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8HQN
 
 | | Activation mechanism of GPR132 by 9(S)-HODE | | Descriptor: | (9S,10E,12Z)-9-hydroxyoctadeca-10,12-dienoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
8HQM
 
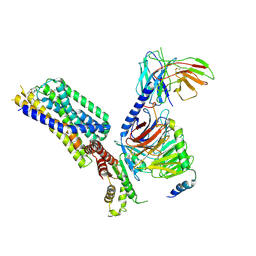 | | Activation mechanism of GPR132 by NPGLY | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
8HQE
 
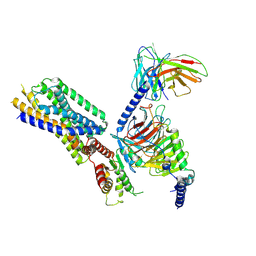 | | Cryo-EM structure of the apo-GPR132-Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
8HVI
 
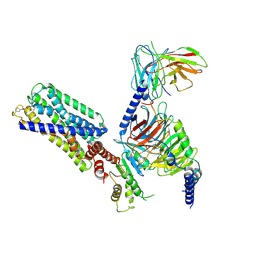 | | Activation mechanism of GPR132 by compound NOX-6-7 | | Descriptor: | 3-methyl-5-[(4-oxidanylidene-4-phenyl-butanoyl)amino]-1-benzofuran-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-26 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
4Q2C
 
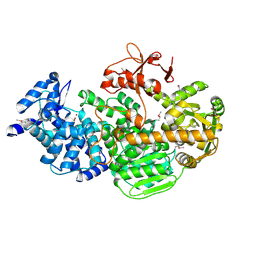 | | Crystal structure of CRISPR-associated protein | | Descriptor: | CRISPR-associated helicase Cas3, NICKEL (II) ION | | Authors: | Gong, B, Shin, M, Sun, J, van der Oost, J, Kim, J.-S. | | Deposit date: | 2014-04-07 | | Release date: | 2014-11-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular insights into DNA interference by CRISPR-associated nuclease-helicase Cas3.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4Q2D
 
 | | Crystal Structure of CRISPR-Associated protein in complex with 2'-Deoxyadenosine 5'-Triphosphate | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CRISPR-associated helicase Cas3, MAGNESIUM ION, ... | | Authors: | Gong, B, Shin, M, Sun, J, van der Oost, J, Kim, J.-S. | | Deposit date: | 2014-04-07 | | Release date: | 2014-11-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.771 Å) | | Cite: | Molecular insights into DNA interference by CRISPR-associated nuclease-helicase Cas3.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NFT
 
 | | Crystal structure of human lnkH2B-h2A.Z-Anp32e | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member E, Histone H2B type 2-E, Histone H2A.Z | | Authors: | Shan, S, Pan, L, Mao, Z, Wang, W, Sun, J, Dong, Q, Liang, X, Ding, X, Chen, S, Dai, L, Zhang, Z, Zhu, B, Zhou, Z. | | Deposit date: | 2013-11-01 | | Release date: | 2014-04-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Anp32e, a higher eukaryotic histone chaperone directs preferential recognition for H2A.Z
Cell Res., 24, 2014
|
|
4QBW
 
 | | The second sphere residue T263 is important for function and activity of PTP1B through modulating WPD loop | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xiao, P, Wang, X, Wang, H.M, Fu, X.L, Cui, F.A, Yu, X, Bi, W.X, Sun, J.P. | | Deposit date: | 2014-05-08 | | Release date: | 2015-02-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.912 Å) | | Cite: | The second-sphere residue T263 is important for the function and catalytic activity of PTP1B via interaction with the WPD-loop
Int.J.Biochem.Cell Biol., 57, 2014
|
|
3U9G
 
 | | Crystal structure of the Zinc finger antiviral protein | | Descriptor: | ZINC ION, Zinc finger CCCH-type antiviral protein 1 | | Authors: | Chen, S, Xu, Y, Zhang, K, Wang, X, Sun, J, Gao, G, Liu, Y. | | Deposit date: | 2011-10-18 | | Release date: | 2012-03-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of N-terminal domain of ZAP indicates how a zinc-finger protein recognizes complex RNA.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4QAP
 
 | | The second sphere residue T263 is important for function and activity of PTP1B through modulating WPD loop | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xiao, P, Wang, X, Wang, H.M, Fu, X.L, Cui, F.A, Yu, X, Bi, W.X, Sun, J.P. | | Deposit date: | 2014-05-05 | | Release date: | 2015-02-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | The second-sphere residue T263 is important for the function and catalytic activity of PTP1B via interaction with the WPD-loop
Int.J.Biochem.Cell Biol., 57, 2014
|
|
4GKF
 
 | | Crystal structure and characterization of Cmr5 protein from Pyrococcus furiosus | | Descriptor: | CRISPR system Cmr subunit Cmr5 | | Authors: | Park, J, Sun, J, Park, S, Hwang, H, Park, M, Shin, M.S. | | Deposit date: | 2012-08-11 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Cmr5 from Pyrococcus furiosus and its functional implications
Febs Lett., 587, 2013
|
|
4RAG
 
 | | CRYSTAL STRUCTURE of PPC2A-D38K | | Descriptor: | MANGANESE (II) ION, Protein phosphatase 1A | | Authors: | Pan, C, Tang, J.Y, Xu, Y.F, Xiao, P, Liu, H.D, Wang, H.A, Wang, W.B, Meng, F.G, Yu, X, Sun, J.P. | | Deposit date: | 2014-09-10 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The catalytic role of the M2 metal ion in PP2C alpha
Sci Rep, 5, 2015
|
|
4RAF
 
 | | Crystal structure of PP2Ca-D38A | | Descriptor: | MANGANESE (II) ION, Protein phosphatase 1A | | Authors: | Pan, C, Tang, J.Y, Xu, Y.F, Xiao, P, Liu, H.D, Wang, H.A, Wang, W.B, Meng, F.G, Yu, X, Sun, J.P. | | Deposit date: | 2014-09-10 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The catalytic role of the M2 metal ion in PP2C alpha
Sci Rep, 5, 2015
|
|
4GFU
 
 | | PTPN18 in complex with HER2-pY1248 phosphor-peptides | | Descriptor: | HER2-pY1248 phosphor-peptide, Tyrosine-protein phosphatase non-receptor type 18 | | Authors: | Wang, H.M, Yang, F, Du, Y.J, Yang, D.X, Zhang, Y, Yu, X, Sun, J.P. | | Deposit date: | 2012-08-04 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PTPN18-HER2 peptides
To be Published
|
|
4QLP
 
 | | Atomic structure of tuberculosis necrotizing toxin (TNT) complexed with its immunity factor IFT | | Descriptor: | Alanine and proline rich protein, tuberculosis necrotizing toxin (TNT), immunity factor IFT | | Authors: | Cingolani, G, Lokareddy, R.K, Sun, J, Siroy, A, Speer, A, Doornbos, K.S, Niederweis, M. | | Deposit date: | 2014-06-12 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The tuberculosis necrotizing toxin kills macrophages by hydrolyzing NAD.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5ZJL
 
 | |
6A67
 
 | | Crystal structure of influenza A virus H5 hemagglutinin globular head in complex with the Fab of antibody FLD21.140 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLD21.140 Heavy Chain, FLD21.140 Light Chain, ... | | Authors: | Wang, P, Zuo, Y, Sun, J, Zhang, L, Wang, X. | | Deposit date: | 2018-06-26 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Complementary recognition of the receptor-binding site of highly pathogenic H5N1 influenza viruses by two human neutralizing antibodies.
J. Biol. Chem., 293, 2018
|
|
4EFO
 
 | | Crystal structure of the ubiquitin-like domain of human TBK1 | | Descriptor: | Serine/threonine-protein kinase TBK1 | | Authors: | Li, J, Li, J, Miyahira, A, Sun, J, Liu, Y, Cheng, G, Liang, H. | | Deposit date: | 2012-03-30 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | Crystal structure of the ubiquitin-like domain of human TBK1.
Protein Cell, 3, 2012
|
|
4GFV
 
 | | PTPN18 in complex with HER2-pY1196 phosphor-peptides | | Descriptor: | HER2-pY1196 phosphor-peptide, Tyrosine-protein phosphatase non-receptor type 18 | | Authors: | Wang, H.M, Yang, F, Du, Y.J, Yang, D.X, Zhang, Y, Yu, X, Sun, J.P. | | Deposit date: | 2012-08-04 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | PTPN18-HER2 peptides
To be Published
|
|
4HA7
 
 | |
4HA9
 
 | |
