7EKP
 
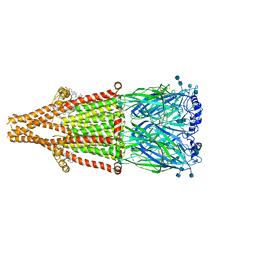 | | human alpha 7 nicotinic acetylcholine receptor bound to EVP-6124 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-chloro-N-(quinuclidin-3-yl)benzo[b]thiophene-2-carboxamide, ... | | Authors: | Liu, S, Zhao, Y, Sun, D, Tian, C. | | Deposit date: | 2021-04-06 | | Release date: | 2021-05-19 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of human alpha 7 nicotinic acetylcholine receptor activation.
Cell Res., 31, 2021
|
|
2AO5
 
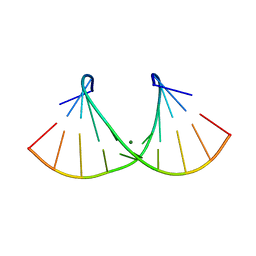 | | Crystal structure of an RNA duplex r(GGCGBrUGCGCU)2 with terminal and internal tandem G-U base pairs | | Descriptor: | 5'-R(*GP*GP*CP*GP*(5BU)P*GP*CP*GP*CP*U)-3', MAGNESIUM ION | | Authors: | Utsunomiya, R, Suto, K, Balasundaresan, D, Fukamizu, A, Kumar, P.K, Mizuno, H. | | Deposit date: | 2005-08-12 | | Release date: | 2006-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of an RNA duplex r(GGCGBrUGCGCU)2 with terminal and internal tandem G.U base pairs.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1TH3
 
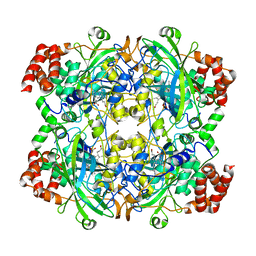 | | Crystal structure of NADPH depleted bovine live catalase complexed with cyanide | | Descriptor: | CYANIDE ION, Catalase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugadev, R, Balasundaresan, D, Ponnuswamy, M.N, Kumaran, D, Swaminathan, S, Sekar, K. | | Deposit date: | 2004-06-01 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of bovine liver catalase
TO BE PUBLISHED
|
|
1TH2
 
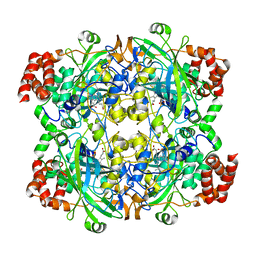 | | crystal structure of NADPH depleted bovine liver catalase complexed with azide | | Descriptor: | AZIDE ION, Catalase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugadev, R, Balasundaresan, D, Ponnuswamy, M.N, Kumaran, D, Swaminathan, S, Sekar, K. | | Deposit date: | 2004-06-01 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of bovine liver catalase
TO BE PUBLISHED
|
|
1TGU
 
 | | The crystal structure of bovine liver catalase without NADPH | | Descriptor: | Catalase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugadev, R, Balasundaresan, D, Ponnuswamy, M.N, Kumaran, D, Swaminathan, S, Sekar, K. | | Deposit date: | 2004-05-31 | | Release date: | 2005-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of bovine liver catalase
TO BE PUBLISHED
|
|
3BOY
 
 | | Crystal structure of the HutP antitermination complex bound to the HUT mRNA | | Descriptor: | 5'-R(*UP*UP*UP*AP*GP*UP*UP*UP*UP*UP*AP*GP*UP*UP*UP*UP*UP*AP*GP*UP*UP*U)-3', HISTIDINE, Hut operon positive regulatory protein, ... | | Authors: | Kumarevel, T.S, Balasundaresan, D, Jeyakanthan, J, Shinkai, A, Yokoyama, S, Kumar, P.K.R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-12-18 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of HutP complexed with the 55-mer RNA
To be Published
|
|
2Y69
 
 | | Bovine heart cytochrome c oxidase re-refined with molecular oxygen | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, CHOLIC ACID, ... | | Authors: | Kaila, V.R.I, Oksanen, E, Goldman, A, Verkhovsky, M.I, Sundholm, D, Wikstrom, M. | | Deposit date: | 2011-01-20 | | Release date: | 2011-02-23 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Combined Quantum Chemical and Crystallographic Study on the Oxidized Binuclear Center of Cytochrome C Oxidase.
Biochim.Biophys.Acta, 1807, 2011
|
|
2LZ5
 
 | | Solution structure of a Novel Alpha-Conotoxin TxIB | | Descriptor: | Conotoxin_TxIB | | Authors: | Luo, S, Zhangsun, D, Wu, Y, Zhu, X, Hu, Y, McIntyre, M, Christensen, S, Akcan, M, Craik, D, McIntosh, J. | | Deposit date: | 2012-09-23 | | Release date: | 2012-12-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Characterization of a novel alpha-conotoxin from conus textile that selectively targets alpha6/alpha3beta2beta3 nicotinic acetylcholine receptors.
J.Biol.Chem., 288, 2013
|
|
2ZFA
 
 | | Structure of Lactate Oxidase at pH4.5 from AEROCOCCUS VIRIDANS | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, Lactate oxidase | | Authors: | Furuichi, M, Balasundaresan, D, Suzuki, N, Yoshida, Y, Minagawa, H, Kaneko, H, Waga, I, Kumar, P.K.R, Mizuno, H. | | Deposit date: | 2007-12-26 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray structures of Aerococcus viridans lactate oxidase and its complex with D-lactate at pH 4.5 show an alpha-hydroxyacid oxidation mechanism
J.Mol.Biol., 378, 2008
|
|
2M3I
 
 | | Characterization of a Novel Alpha4/6-Conotoxin TxIC from Conus textile that Potently Blocks alpha3beta4 Nicotinic Acetylcholine Receptors | | Descriptor: | Alpha-conotoxin | | Authors: | Luo, S, Zhangsun, D, Zhu, X, Wu, Y, Hu, Y, Christensen, S, Akcan, M, Craik, D.J, McIntosh, J.M. | | Deposit date: | 2013-01-20 | | Release date: | 2013-12-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Characterization of a Novel alpha-Conotoxin TxID from Conus textile That Potently Blocks Rat alpha 3 beta 4 Nicotinic Acetylcholine Receptors.
J.Med.Chem., 56, 2013
|
|
2MWA
 
 | |
2MWF
 
 | |
2MWB
 
 | | FBP28 WW2 mutant W457F | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Macias, M.J, Scheraga, H, Sunol, D, Todorovski, T. | | Deposit date: | 2014-11-03 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding kinetics of WW domains with the united residue force field for bridging microscopic motions and experimental measurements.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2MWD
 
 | |
2MW9
 
 | |
2MWE
 
 | | NMR structure of FBP28 WW2 mutant Y438R, L453A DNDC | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Macias, M.J, Scheraga, H, Sunol, D, Todorovski, T. | | Deposit date: | 2014-11-04 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding kinetics of WW domains with the united residue force field for bridging microscopic motions and experimental measurements.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2NLI
 
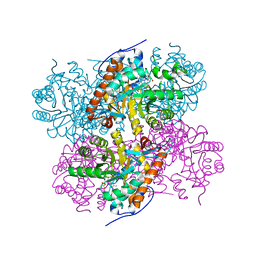 | | Crystal Structure of the complex between L-lactate oxidase and a substrate analogue at 1.59 angstrom resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, HYDROGEN PEROXIDE, LACTIC ACID, ... | | Authors: | Furuichi, M, Suzuki, N, Balasundaresan, D, Yoshida, Y, Minagawa, H, Watanabe, Y, Kaneko, H, Waga, I, Kumar, P.K.R, Mizuno, H. | | Deposit date: | 2006-10-20 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | X-ray structures of Aerococcus viridans lactate oxidase and its complex with D-lactate at pH 4.5 show an alpha-hydroxyacid oxidation mechanism
J.Mol.Biol., 378, 2008
|
|
