5MYX
 
 | |
3CBG
 
 | | Functional and Structural Characterization of a Cationdependent O-Methyltransferase from the Cyanobacterium Synechocystis Sp. Strain PCC 6803 | | Descriptor: | (2E)-3-(3-hydroxy-4-methoxyphenyl)prop-2-enoic acid, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, MAGNESIUM ION, ... | | Authors: | Kopycki, J.G, Neumann, P, Stubbs, M.T. | | Deposit date: | 2008-02-22 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and Structural Characterization of a Cation-dependent O-Methyltransferase from the Cyanobacterium Synechocystis sp. Strain PCC 6803
J.Biol.Chem., 283, 2008
|
|
3LN9
 
 | | Crystal structure of the fibril-specific B10 antibody fragment | | Descriptor: | CITRATE ANION, GLYCEROL, Immunoglobulin heavy chain antibody variable domain B10, ... | | Authors: | Parthier, C, Morgado, I, Stubbs, M.T, Faendrich, M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Amyloid Fibril Recognition with the Conformational B10 Antibody Fragment Depends on Electrostatic Interactions.
J.Mol.Biol., 2010
|
|
1MD9
 
 | | CRYSTAL STRUCTURE OF DhbE IN COMPLEX WITH DHB AND AMP | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, 2,3-dihydroxybenzoate-AMP ligase, ADENOSINE MONOPHOSPHATE | | Authors: | May, J.J, Kessler, N, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2002-08-07 | | Release date: | 2002-09-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4F9U
 
 | | Structure of glycosylated glutaminyl cyclase from Drosophila melanogaster | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CG32412, ... | | Authors: | Kolenko, P, Koch, B, Ruiz-Carilo, D, Stubbs, M.T. | | Deposit date: | 2012-05-21 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Glutaminyl Cyclases (QCs) from Drosophila melanogaster Reveal Active Site Conservation between Insect and Mammalian QCs.
Biochemistry, 51, 2012
|
|
4F9V
 
 | | Structure of C113A/C136A mutant variant of glycosylated glutaminyl cyclase from Drosophila melanogaster | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, CG32412, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kolenko, P, Koch, B, Ruiz-Carilo, D, Stubbs, M.T. | | Deposit date: | 2012-05-21 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Glutaminyl Cyclases (QCs) from Drosophila melanogaster Reveal Active Site Conservation between Insect and Mammalian QCs.
Biochemistry, 51, 2012
|
|
6QRO
 
 | | Crystal structure of Tannerella forsythia glutaminyl cyclase | | Descriptor: | Glutamine cyclotransferase, SULFATE ION, ZINC ION | | Authors: | Linnert, M, Piechotta, A, Parthier, C, Taudte, N, Kolenko, P, Rahfeld, J, Potempa, J, Stubbs, M.T. | | Deposit date: | 2019-02-19 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mammalian-like type II glutaminyl cyclases in Porphyromonas gingivalis and other oral pathogenic bacteria as targets for treatment of periodontitis.
J.Biol.Chem., 296, 2021
|
|
6QQL
 
 | | Crystal structure of Porphyromonas gingivalis glutaminyl cyclase | | Descriptor: | Glutamine cyclotransferase, ZINC ION | | Authors: | Linnert, M, Piechotta, A, Parthier, C, Taudte, N, Kolenko, P, Rahfeld, J, Potempa, J, Stubbs, M.T. | | Deposit date: | 2019-02-18 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.814 Å) | | Cite: | Mammalian-like type II glutaminyl cyclases in Porphyromonas gingivalis and other oral pathogenic bacteria as targets for treatment of periodontitis.
J.Biol.Chem., 296, 2021
|
|
4WHE
 
 | | Crystal structure of E. coli phage shock protein A (PspA 1-144) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Phage shock protein A | | Authors: | Parthier, C, Schoepfel, M, Stubbs, M.T, Osadnik, H, Brueser, T. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | PspF-binding domain PspA1-144 and the PspAF complex: New insights into the coiled-coil-dependent regulation of AAA+ proteins.
Mol.Microbiol., 98, 2015
|
|
3UWI
 
 | | Bovine trypsin variant X(tripleGlu217Phe227) in complex with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cationic trypsin, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-12-02 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
3UPE
 
 | | Bovine trypsin variant X(triplePhe227) in complex with small molecule inhibitor | | Descriptor: | 1-[(7-carbamimidoylnaphthalen-2-yl)methyl]-6-({1-[(1Z)-ethanimidoyl]piperidin-4-yl}oxy)-2-(propan-2-yl)-1H-indole-4-carboxylic acid, CALCIUM ION, Cationic trypsin, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-11-18 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
3UQV
 
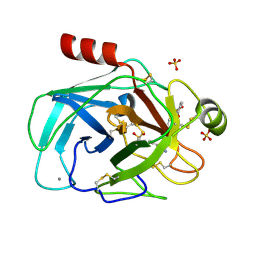 | | Bovine trypsin variant X(triplePhe227) in complex with small molecule inhibitor | | Descriptor: | 3-(3-carbamimidoylphenyl)-N-(2'-sulfamoylbiphenyl-4-yl)-1,2-oxazole-4-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-11-21 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
3UY9
 
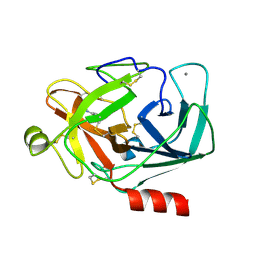 | | Bovine trypsin variant X(tripleGlu217Phe227) in complex with small molecule inhibitor | | Descriptor: | BENZAMIDINE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-12-06 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
3UQO
 
 | | Bovine trypsin variant X(triplePhe227) in complex with small molecule inhibitor | | Descriptor: | 3-(3-carbamimidoylphenyl)-N-(2'-sulfamoylbiphenyl-4-yl)-1,2-oxazole-4-carboxamide, CALCIUM ION, Cationic trypsin | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-11-21 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
6GV1
 
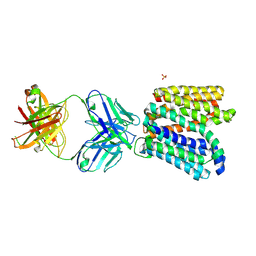 | | Crystal structure of E.coli Multidrug/H+ antiporter MdfA in outward open conformation with bound Fab fragment | | Descriptor: | Fab fragment YN1074 heavy chain, Fab fragment YN1074 light chain, Major Facilitator Superfamily multidrug/H+ antiporter MdfA from E.coli, ... | | Authors: | Nagarathinam, K, Parthier, C, Stubbs, M.T, Tanabe, M. | | Deposit date: | 2018-06-20 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Outward open conformation of a Major Facilitator Superfamily multidrug/H+antiporter provides insights into switching mechanism.
Nat Commun, 9, 2018
|
|
1BTH
 
 | |
8PE4
 
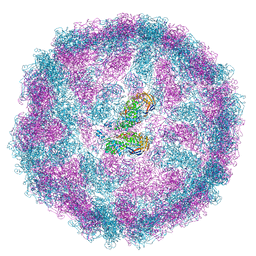 | |
1A5I
 
 | |
2BMA
 
 | | The crystal structure of Plasmodium falciparum glutamate dehydrogenase, a putative target for novel antimalarial drugs | | Descriptor: | GLUTAMATE DEHYDROGENASE (NADP+) | | Authors: | Werner, C, Stubbs, M.T, Krauth-Siege, R.L, Klebe, G. | | Deposit date: | 2005-03-10 | | Release date: | 2005-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of Plasmodium Falciparum Glutamate Dehydrogenase, a Putative Target for Novel Antimalarial Drugs
J.Mol.Biol., 349, 2005
|
|
6SHT
 
 | | Molecular structure of mouse apoferritin resolved at 2.7 Angstroms with the Glacios cryo-microscope | | Descriptor: | FE (III) ION, Ferritin heavy chain, MAGNESIUM ION | | Authors: | Hamdi, F, Tueting, C, Semchonok, D, Kyrilis, F, Meister, A, Skalidis, I, Schmidt, L, Parthier, C, Stubbs, M.T, Kastritis, P.L. | | Deposit date: | 2019-08-08 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | 2.7 angstrom cryo-EM structure of vitrified M. musculus H-chain apoferritin from a compact 200 keV cryo-microscope.
Plos One, 15, 2020
|
|
8A5T
 
 | |
3C3Y
 
 | | Crystal Structure of PFOMT, Phenylpropanoid and Flavonoid O-methyltransferase from M. crystallinum | | Descriptor: | CALCIUM ION, O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kopycki, J.G, Rauh, D, Neumann, P, Stubbs, M.T. | | Deposit date: | 2008-01-29 | | Release date: | 2008-04-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.371 Å) | | Cite: | Biochemical and Structural Analysis of Substrate Promiscuity in Plant Mg(2+)-Dependent O-Methyltransferases
J.Mol.Biol., 378, 2008
|
|
1A5H
 
 | |
5MY4
 
 | |
5MYK
 
 | |
