6WZZ
 
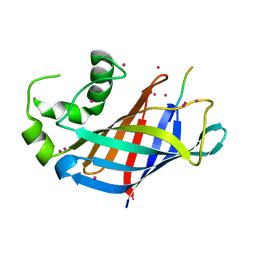 | | GID4 in complex with VGLWKS peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, UNKNOWN ATOM OR ION, VGLWKS peptide | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-14 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of nonproline N-terminal residues by the Pro/N-degron pathway.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5KH9
 
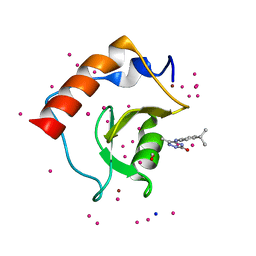 | | Crystal structure of a low occupancy fragment candidate (5-[(4-Isopropylphenyl)amino]-6-methyl-1,2,4-triazin-3(2H)-one) bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 6-methyl-5-[(4-propan-2-ylphenyl)amino]-2~{H}-1,2,4-triazin-3-one, FORMIC ACID, Histone deacetylase 6, ... | | Authors: | Harding, R.J, Tempel, W, Ravichandran, M, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, Schapira, M, Bountra, C, Edwards, A.M, von Delft, F, Santhakumar, V, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
7LMT
 
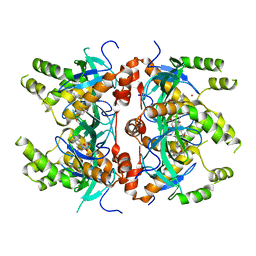 | | Histone-lysine N-methyltransferase NSD2-PWWP1 with compound MRT10241866a | | Descriptor: | Histone-lysine N-methyltransferase NSD2, UNKNOWN ATOM OR ION, ~{N}-cyclopropyl-3-oxidanylidene-~{N}-(thiophen-2-ylmethyl)-4~{H}-1,4-benzoxazine-7-carboxamide | | Authors: | Lei, M, Freitas, R.F, Dong, A, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Histone-lysine N-methyltransferase NSD2-PWWP1 with compound MRT10241866a
To Be Published
|
|
6Q8K
 
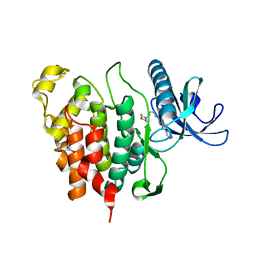 | | CLK1 with bound pyridoquinazoline | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein kinase CLK1, ~{N}2-(3-morpholin-4-ylpropyl)pyrido[3,4-g]quinazoline-2,10-diamine | | Authors: | Schroeder, M, Tazarki, H, Zeinyeh, W, Esvan, Y.J, Khiari, J, Joesselin, B, Bach, S, Ruchaud, S, Anizon, F, Giraud, F, Moreau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | New pyrido[3,4-g]quinazoline derivatives as CLK1 and DYRK1A inhibitors: synthesis, biological evaluation and binding mode analysis.
Eur J Med Chem, 166, 2019
|
|
7MDN
 
 | | Histone-lysine N-methyltransferase NSD2-PWWP1 with compound MRT10241866a | | Descriptor: | Histone-lysine N-methyltransferase NSD2, UNKNOWN ATOM OR ION, ~{N}-cyclopropyl-3-oxidanylidene-~{N}-(thiophen-2-ylmethyl)-4~{H}-1,4-benzoxazine-7-carboxamide | | Authors: | Lei, M, Freitas, R.F, Dong, A, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-05 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | A chemical probe targeting the PWWP domain alters NSD2 nucleolar localization.
Nat.Chem.Biol., 18, 2022
|
|
5LVR
 
 | | Crystal structure of human PCAF bromodomain in complex with compound-E (CPD-E) | | Descriptor: | 1,2-ETHANEDIOL, 5-methyl-2-phenyl-1,2,3-triazole-4-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, Filippakopoulos, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hopkins, A.L, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of New Bromodomain Scaffolds by Biosensor Fragment Screening.
ACS Med Chem Lett, 7, 2016
|
|
6Y6V
 
 | | p38a bound with MCP-81 | | Descriptor: | 5-azanyl-~{N}-[[4-[[5-~{tert}-butyl-2-(4-methylphenyl)pyrazol-3-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Schroeder, M, Roehm, S, Knapp, S, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-27 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
5LVQ
 
 | | Crystal structure of human PCAF bromodomain in complex with compound-D (CPD-D), N-methyl-2-(tetrahydro-2H-pyran-4-yloxy)benzamide | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Histone acetyltransferase KAT2B, ... | | Authors: | Chaikuad, A, Filippakopoulos, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hopkins, A.L, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of New Bromodomain Scaffolds by Biosensor Fragment Screening.
ACS Med Chem Lett, 7, 2016
|
|
5BT4
 
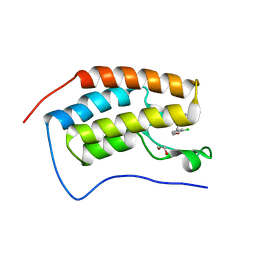 | | Crystal structure of BRD4 first bromodomain in complex with SGC-CBP30 chemical probe | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(3-chloro-4-methoxyphenyl)ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(2S)-2-(morpholin-4-yl)propyl]-1H-benzimidazole, Bromodomain-containing protein 4 | | Authors: | Tallant, C, Hay, D, Krojer, T, Nunez-Alonso, G, Picaud, S, Newman, J.A, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of BRD4 first bromodomain in complex with a 3,5-dimethylisoxazol ligand
To Be Published
|
|
5BT5
 
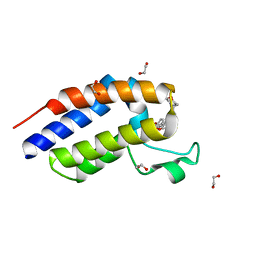 | | Crystal structure of BRD2 second bromodomain in complex with SGC-CBP30 chemical probe | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(3-chloro-4-methoxyphenyl)ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(2S)-2-(morpholin-4-yl)propyl]-1H-benzimidazole, Bromodomain-containing protein 2 | | Authors: | Tallant, C, Hay, D, Krojer, T, Nunez-Alonso, G, Picaud, S, Newman, J.A, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of BRD2 second bromodomain in complex with a 3,5-dimethylisoxazol ligand
To Be Published
|
|
5BT3
 
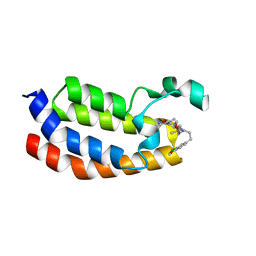 | | Crystal structure of EP300 bromodomain in complex with SGC-CBP30 chemical probe | | Descriptor: | 2-[2-(3-chloro-4-methoxyphenyl)ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(2S)-2-(morpholin-4-yl)propyl]-1H-benzimidazole, Histone acetyltransferase p300, ISOPROPYL ALCOHOL | | Authors: | Tallant, C, Hay, D, Krojer, T, Nunez-Alonso, G, Picaud, S, Newman, J.A, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal structure of EP300 bromodomain in complex with a 3,5-dimethylisoxazol ligand
To Be Published
|
|
3B7O
 
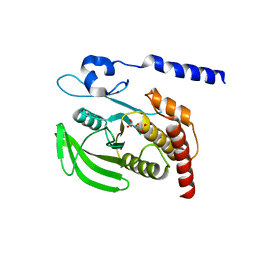 | | Crystal structure of the human tyrosine phosphatase SHP2 (PTPN11) with an accessible active site | | Descriptor: | D-MALATE, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Ugochukwu, E, Barr, A, Patel, A, King, O, Niesen, F, Salah, E, Savitsky, P, Pilka, E.S, Elkins, J, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-31 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
6YG5
 
 | |
6YG0
 
 | |
6YG6
 
 | | Crystal structure of MKK7 (MAP2K7) covalently bound with type-II inhibitor TL10-105 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity mitogen-activated protein kinase kinase 7, ~{N}-[4-[(4-ethylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl]-4-methyl-3-[2-[[(3~{S})-1-propanoylpyrrolidin-3-yl]amino]pyrimidin-4-yl]oxy-benzamide | | Authors: | Chaikuad, A, Tan, L, Wang, J, Liang, Y, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-27 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Catalytic Domain Plasticity of MKK7 Reveals Structural Mechanisms of Allosteric Activation and Diverse Targeting Opportunities.
Cell Chem Biol, 27, 2020
|
|
5II1
 
 | | Crystal Structure of the fifth bromodomain of human polybromo (PB1) in complex with 1-methylisochromeno[3,4-c]pyrazol-5(3H)-one | | Descriptor: | 1-methyl[2]benzopyrano[3,4-c]pyrazol-5(3H)-one, Protein polybromo-1 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Myrianthopoulos, V, Mikros, E, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-01 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Discovery and Optimization of a Selective Ligand for the Switch/Sucrose Nonfermenting-Related Bromodomains of Polybromo Protein-1 by the Use of Virtual Screening and Hydration Analysis.
J.Med.Chem., 59, 2016
|
|
6YZ4
 
 | | Crystal structure of MKK7 (MAP2K7) with ibrutinib bound at allosteric site | | Descriptor: | 1,2-ETHANEDIOL, 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-06 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic Domain Plasticity of MKK7 Reveals Structural Mechanisms of Allosteric Activation and Diverse Targeting Opportunities.
Cell Chem Biol, 27, 2020
|
|
6WNX
 
 | | FBXW11-SKP1 in complex with a pSer33/pSer37 Beta-Catenin peptide | | Descriptor: | Catenin beta-1, F-box/WD repeat-containing protein 11, GLYCEROL, ... | | Authors: | Ivanochko, D, Edwards, A.M, Bountra, C, Arrowsmith, C.H, Boettcher, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-23 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | FBXW11-SKP1 in complex with a pSer33/pSer37 Beta-Catenin peptide
To Be Published
|
|
5KH3
 
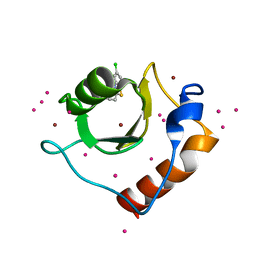 | | Crystal structure of fragment (3-(5-Chloro-1,3-benzothiazol-2-yl)propanoic acid) bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(5-chloranyl-1,3-benzothiazol-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Dong, A, Ravichandran, M, Ferreira de Freitas, R, Schapira, M, Bountra, C, Edwards, A.M, Santhakumar, V, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
5KH7
 
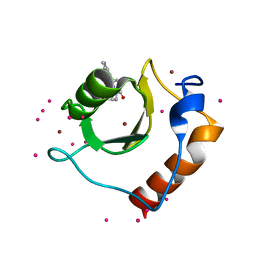 | | Crystal structure of fragment (3-[6-Oxo-3-(3-pyridinyl)-1(6H)-pyridazinyl]propanoic acid) bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(6-oxidanylidene-3-pyridin-3-yl-pyridazin-1-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Walker, J, Ravichandran, M, Ferreira de Freitas, R, Schapira, M, Bountra, C, Edwards, A.M, Santhakumar, V, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
6XCG
 
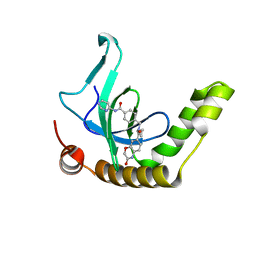 | | Histone-lysine N-methyltransferase NSD2-PWWP1 with compound UNC6934 | | Descriptor: | Histone-lysine N-methyltransferase NSD2, N-cyclopropyl-3-oxo-N-({4-[(pyrimidin-4-yl)carbamoyl]phenyl}methyl)-3,4-dihydro-2H-1,4-benzoxazine-7-carboxamide, UNKNOWN ATOM OR ION | | Authors: | Zhou, M.Q, Dong, A, Ingerman, L.A, Hanley, R.P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A chemical probe targeting the PWWP domain alters NSD2 nucleolar localization.
Nat.Chem.Biol., 18, 2022
|
|
6TCX
 
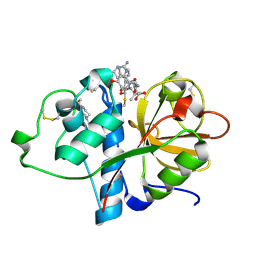 | | Papain bound to a natural cysteine protease inhibitor from Streptomyces mobaraensis | | Descriptor: | (2~{R})-2-[[(1~{S})-1-[(6~{S})-2-azanyl-1,4,5,6-tetrahydropyrimidin-6-yl]-2-[[(2~{S})-3-methyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanyl-3-phenyl-propan-2-yl]amino]butan-2-yl]amino]-2-oxidanylidene-ethyl]carbamoylamino]-3-(4-hydroxyphenyl)propanoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Papain | | Authors: | Kraemer, A, Juettner, N.E, Fuchsbauer, H.-L, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-06 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Decoding the Papain Inhibitor from Streptomyces mobaraensis as Being Hydroxylated Chymostatin Derivatives: Purification, Structure Analysis, and Putative Biosynthetic Pathway.
J.Nat.Prod., 83, 2020
|
|
6D1T
 
 | | Complex of MBD1-MBD and methylated DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain protein 1, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Xu, C, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-04-12 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Complex of MBD1-MBD and methylated DNA
to be published
|
|
2Z5D
 
 | | Human ubiquitin-conjugating enzyme E2 H | | Descriptor: | SODIUM ION, Ubiquitin-conjugating enzyme E2 H | | Authors: | Bochkarev, A, Cui, H, Walker, J.R, Newman, E.M, Mackenzie, F, Battaile, K.P, Sundstrom, M, Arrowsmith, C, Edwards, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-06 | | Release date: | 2007-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
MOL.CELL PROTEOMICS, 11, 2012
|
|
2ZFY
 
 | | Crystal structure of human Otubain 1 | | Descriptor: | Ubiquitin thioesterase OTUB1 | | Authors: | Akutsu, M, Walker, J.R, Li, Y, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-16 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis and specificity of human otubain 1-mediated deubiquitination.
Biochem.J., 418, 2009
|
|
