4JLT
 
 | | Crystal structure of P450 2B4(H226Y) in complex with paroxetine | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, GLYCEROL, ... | | Authors: | Shah, M.B, Pascual, J, Stout, C.D, Halpert, J.R. | | Deposit date: | 2013-03-13 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A Structural Snapshot of CYP2B4 in Complex with Paroxetine Provides Insights into Ligand Binding and Clusters of Conformational States.
J.Pharmacol.Exp.Ther., 346, 2013
|
|
4K4R
 
 | |
4K4P
 
 | | TL-3 inhibited Trp6Ala HIV Protease | | Descriptor: | HIV-1 protease, NITRATE ION, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Tiefenbrunn, T, Stout, C.D. | | Deposit date: | 2013-04-12 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystallographic Fragment-Based Drug Discovery: Use of a Brominated Fragment Library Targeting HIV Protease.
Chem.Biol.Drug Des., 83, 2014
|
|
3MDT
 
 | | Voriconazole complex of Cytochrome P450 46A1 | | Descriptor: | Cholesterol 24-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE, Voriconazole | | Authors: | Mast, N, Charvet, C, Pikuleva, I, Stout, C.D. | | Deposit date: | 2010-03-30 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of drug binding to CYP46A1, an enzyme that controls cholesterol turnover in the brain.
J.Biol.Chem., 285, 2010
|
|
4K4Q
 
 | | TL-3 inhibited Trp6Ala HIV Protease with 3-bromo-2,6-dimethoxybenzoic acid bound in flap site | | Descriptor: | 3-bromo-2,6-dimethoxybenzoic acid, BETA-MERCAPTOETHANOL, DIMETHYL SULFOXIDE, ... | | Authors: | Tiefenbrunn, T, Stout, C.D. | | Deposit date: | 2013-04-12 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Fragment-Based Drug Discovery: Use of a Brominated Fragment Library Targeting HIV Protease.
Chem.Biol.Drug Des., 83, 2014
|
|
3MDM
 
 | | Thioperamide complex of Cytochrome P450 46A1 | | Descriptor: | Cholesterol 24-hydroxylase, N-cyclohexyl-4-(1H-imidazol-5-yl)piperidine-1-carbothioamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mast, N, Charvet, C, Pikuleva, I, Stout, C.D. | | Deposit date: | 2010-03-30 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of drug binding to CYP46A1, an enzyme that controls cholesterol turnover in the brain.
J.Biol.Chem., 285, 2010
|
|
3MDV
 
 | | Clotrimazole complex of Cytochrome P450 46A1 | | Descriptor: | 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, Cholesterol 24-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mast, N, Charvet, C, Pikuleva, I, Stout, C.D. | | Deposit date: | 2010-03-30 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of drug binding to CYP46A1, an enzyme that controls cholesterol turnover in the brain.
J.Biol.Chem., 285, 2010
|
|
3MDR
 
 | | Tranylcypromine complex of Cytochrome P450 46A1 | | Descriptor: | (1R,2S)-2-phenylcyclopropanamine, Cholesterol 24-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mast, N, Charvet, C, Pikuleva, I, Stout, C.D. | | Deposit date: | 2010-03-30 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of drug binding to CYP46A1, an enzyme that controls cholesterol turnover in the brain.
J.Biol.Chem., 285, 2010
|
|
4J1T
 
 | | Crystal structure of Thermus thermophilus transhydrogenase heterotrimeric complex of the Alpha1 subunit dimer with the NADP binding domain (domain III) of the Beta subunit in P2(1) | | Descriptor: | GLYCEROL, NAD(P) transhydrogenase subunit beta, NAD/NADP transhydrogenase alpha subunit 1, ... | | Authors: | Yamaguchi, M, Leung, J, Schurig Briccio, L.A, Gennis, R.B, Stout, C.D. | | Deposit date: | 2013-02-02 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure analysis of Thermus thermophilus transhydrogenase soluble domains
To be Published
|
|
4G72
 
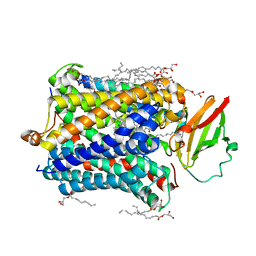 | |
4G7Q
 
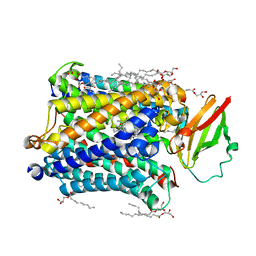 | |
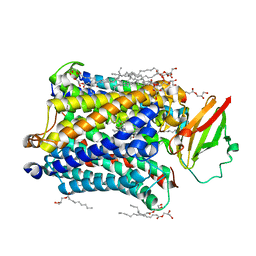
4G71
 
 | |
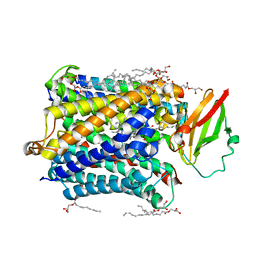
4G7R
 
 | |
4G70
 
 | |
4G7S
 
 | |
4GP4
 
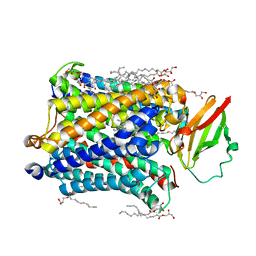 | | Structure of Recombinant Cytochrome ba3 Oxidase mutant Y133F from Thermus thermophilus | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Li, Y, Chen, Y, Stout, C.D. | | Deposit date: | 2012-08-20 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ligand Access to the Active Site in Thermus thermophilusba(3) and Bovine Heart aa(3) Cytochrome Oxidases.
Biochemistry, 52, 2013
|
|
4GP8
 
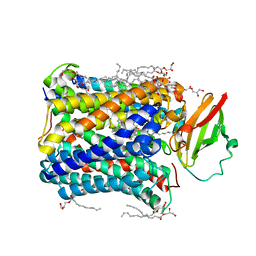 | | Structure of Recombinant Cytochrome ba3 Oxidase mutant Y133W+T231F from Thermus thermophilus | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Li, Y, Chen, Y, Stout, C.D. | | Deposit date: | 2012-08-20 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ligand Access to the Active Site in Thermus thermophilusba(3) and Bovine Heart aa(3) Cytochrome Oxidases.
Biochemistry, 52, 2013
|
|
4GQS
 
 | | Structure of Human Microsomal Cytochrome P450 (CYP) 2C19 | | Descriptor: | (4-hydroxy-3,5-dimethylphenyl)(2-methyl-1-benzofuran-3-yl)methanone, Cytochrome P450 2C19, GLYCEROL, ... | | Authors: | Reynald, R.L, Sansen, S, Stout, C.D, Johnson, E.F. | | Deposit date: | 2012-08-23 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural Characterization of Human Cytochrome P450 2C19: ACTIVE SITE DIFFERENCES BETWEEN P450s 2C8, 2C9, AND 2C19.
J.Biol.Chem., 287, 2012
|
|
4GP5
 
 | | Structure of Recombinant Cytochrome ba3 Oxidase mutant Y133W from Thermus thermophilus | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Li, Y, Chen, Y, Stout, C.D. | | Deposit date: | 2012-08-20 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ligand Access to the Active Site in Thermus thermophilusba(3) and Bovine Heart aa(3) Cytochrome Oxidases.
Biochemistry, 52, 2013
|
|
4HYX
 
 | | Crystal Structure Analysis of the Bacteriorhodopsin in Facial Amphiphile-4 DMPC Bicelle | | Descriptor: | Bacteriorhodopsin, DECANE, GLYCEROL, ... | | Authors: | Lee, S, Stout, C.D, Zhang, Q. | | Deposit date: | 2012-11-14 | | Release date: | 2013-03-20 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HWL
 
 | | Crystal Structure Analysis of the Bacteriorhodopsin in Facial Amphiphile-7 DMPC Bicelle | | Descriptor: | Bacteriorhodopsin, GLYCEROL, HEPTANE, ... | | Authors: | Lee, S, Stout, C.D, Zhang, Q. | | Deposit date: | 2012-11-08 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4I91
 
 | | Crystal Structure of Cytochrome P450 2B6 (Y226H/K262R) in complex with alpha-Pinene. | | Descriptor: | (+)-alpha-Pinene, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B6, ... | | Authors: | Shah, M.B, Stout, C.D, Halpert, J.R. | | Deposit date: | 2012-12-04 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Thermodynamic Basis of (+)-alpha-Pinene Binding to Human Cytochrome P450 2B6.
J.Am.Chem.Soc., 135, 2013
|
|
2Q9F
 
 | | Crystal structure of human cytochrome P450 46A1 in complex with cholesterol-3-sulphate | | Descriptor: | CHOLEST-5-EN-3-YL HYDROGEN SULFATE, Cytochrome P450 46A1, GLYCEROL, ... | | Authors: | White, M.A, Mast, N.V, Johnson, E.F, Stout, C.D, Pikuleva, I.A. | | Deposit date: | 2007-06-12 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of substrate-bound and substrate-free cytochrome P450 46A1, the principal cholesterol hydroxylase in the brain.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2Q9G
 
 | | Crystal structure of human cytochrome P450 46A1 | | Descriptor: | Cytochrome P450 46A1, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | White, M.A, Mast, N.V, Johnson, E.F, Stout, C.D, Pikuleva, I.A. | | Deposit date: | 2007-06-12 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of substrate-bound and substrate-free cytochrome P450 46A1, the principal cholesterol hydroxylase in the brain.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1FTC
 
 | | Y13C MUTANT OF AZOTOBACTER VINELANDII FDI | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Kemper, M.A, Lloyd, S.J, Prasad, G.S, Stout, C.D, Fawcett, S, Armstrong, F.A, Burgess, B.K. | | Deposit date: | 1997-01-08 | | Release date: | 1997-04-01 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Y13C Azotobacter vinelandii ferredoxin I. A designed [Fe-S] ligand motif contains a cysteine persulfide.
J.Biol.Chem., 272, 1997
|
|
