6O3H
 
 | | Icosahedral reconstruction of the thermophilic bacteriophage P74-26 capsid | | Descriptor: | Major head protein, P74-26 Head Decoration Protein | | Authors: | Stone, N.P, Demo, G, Agnello, E, Kelch, B.A. | | Deposit date: | 2019-02-26 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Principles for enhancing virus capsid capacity and stability from a thermophilic virus capsid structure.
Nat Commun, 10, 2019
|
|
6BL5
 
 | | Head decoration protein from the hyperthermophilic phage P74-26 | | Descriptor: | Head decoration protein | | Authors: | Stone, N.P, Hilbert, B.J, Hidalgo, D, Halloran, K.T, Kelch, B.A. | | Deposit date: | 2017-11-09 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A Hyperthermophilic Phage Decoration Protein Suggests Common Evolutionary Origin with Herpesvirus Triplex Proteins and an Anti-CRISPR Protein.
Structure, 26, 2018
|
|
6V1I
 
 | | Cryo-EM reconstruction of the thermophilic bacteriophage P74-26 small terminase- symmetric | | Descriptor: | Small terminase protein | | Authors: | Hayes, J.A, Hilbert, B.J, Gaubitz, C, Stone, N.P, Kelch, B.A. | | Deposit date: | 2019-11-20 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A thermophilic phage uses a small terminase protein with a fixed helix-turn-helix geometry.
J.Biol.Chem., 295, 2020
|
|
6VVO
 
 | | Structure of the human clamp loader (Replication Factor C, RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen, PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Stone, N.P, Kelch, B.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the human clamp loader bound to the sliding clamp: a further twist on AAA+ mechanism
Biorxiv, 2020
|
|
4ZNJ
 
 | | Thermus Phage P74-26 Large Terminase ATPase domain mutant R139A (I 2 3 space group) | | Descriptor: | Phage terminase large subunit, SULFATE ION | | Authors: | Hilbert, B.J, Hayes, J.A, Stone, N.P, Duffy, C.M, Kelch, B.A. | | Deposit date: | 2015-05-04 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.532 Å) | | Cite: | Structure and mechanism of the ATPase that powers viral genome packaging.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZNK
 
 | | Thermus Phage P74-26 Large Terminase ATPase domain from (P 32 2 1 space group) | | Descriptor: | Phage terminase large subunit, SULFATE ION | | Authors: | Hilbert, B.J, Hayes, J.A, Stone, N.P, Duffy, C.M, Kelch, B.A. | | Deposit date: | 2015-05-04 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.931 Å) | | Cite: | Structure and mechanism of the ATPase that powers viral genome packaging.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZNI
 
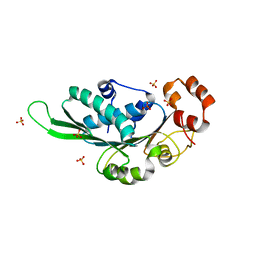 | | Thermus Phage P74-26 Large Terminase ATPase domain (I 2 3 space group) | | Descriptor: | Phage terminase large subunit, SULFATE ION | | Authors: | Hilbert, B.J, Hayes, J.A, Stone, N.P, Duffy, C.M, Sankaran, B, Kelch, B.A. | | Deposit date: | 2015-05-04 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structure and mechanism of the ATPase that powers viral genome packaging.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZNL
 
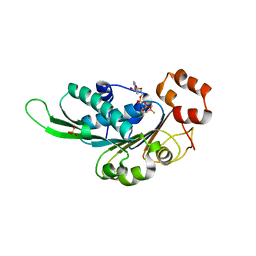 | | Thermus Phage P74-26 Large Terminase ATPase domain bound to ADP Beryllium Fluoride | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Hilbert, B.J, Hayes, J.A, Stone, N.P, Duffy, C.M, Kelch, B.A. | | Deposit date: | 2015-05-04 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.068 Å) | | Cite: | Structure and mechanism of the ATPase that powers viral genome packaging.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5TGE
 
 | |
7TIC
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TIB
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) and primer-template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*AP*GP*AP*CP*AP*CP*TP*AP*CP*GP*AP*GP*TP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*AP*TP*GP*TP*AP*CP*TP*CP*GP*TP*AP*GP*TP*GP*TP*CP*T)-3'), ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TI8
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7THV
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-12 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TID
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) and primer-template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*AP*GP*AP*CP*AP*CP*TP*AP*CP*GP*AP*GP*TP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*AP*TP*GP*TP*AP*CP*TP*CP*GP*TP*AP*GP*TP*GP*TP*CP*T)-3'), ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TKU
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7THJ
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-11 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7MSF
 
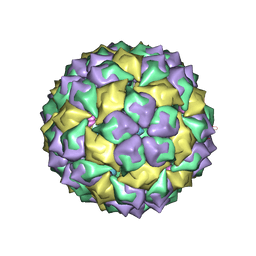 | | MS2 PROTEIN CAPSID/RNA COMPLEX | | Descriptor: | 5'-R(*UP*CP*GP*CP*CP*AP*AP*CP*AP*GP*GP*CP*GP*G)-3', MS2 PROTEIN CAPSID | | Authors: | Rowsell, S, Stonehouse, N.J, Convery, M.A, Adams, C.J, Ellington, A.D, Hirao, I, Peabody, D.S, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 1998-05-20 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of a series of RNA aptamers complexed to the same protein target.
Nat.Struct.Biol., 5, 1998
|
|
3ZL9
 
 | | Crystal structure of the nucleocapsid protein from Schmallenberg virus | | Descriptor: | NUCLEOCAPSID PROTEIN | | Authors: | Ariza, A, Tanner, S.J, Walter, C.T, Dent, K.C, Shepherd, D.A, Wu, W, Matthews, S.V, Hiscox, J.A, Green, T.J, Luo, M, Elliot, R.M, Ashcroft, A.E, Stonehouse, N.J, Ranson, N.A, Barr, J.N, Edwards, T.A. | | Deposit date: | 2013-01-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Nucleocapsid Protein Structures from Orthobunyaviruses Reveal Insight Into Ribonucleoprotein Architecture and RNA Polymerization.
Nucleic Acids Res., 41, 2013
|
|
3ZLA
 
 | | Crystal structure of the nucleocapsid protein from Bunyamwera virus bound to RNA | | Descriptor: | NUCLEOPROTEIN, RNA | | Authors: | Ariza, A, Tanner, S.J, Walter, C.T, Dent, K.C, Shepherd, D.A, Wu, W, Matthews, S.V, Hiscox, J.A, Green, T.J, Luo, M, Elliot, R.M, Ashcroft, A.E, Stonehouse, N.J, Ranson, N.A, Barr, J.N, Edwards, T.A. | | Deposit date: | 2013-01-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Nucleocapsid Protein Structures from Orthobunyaviruses Reveal Insight Into Ribonucleoprotein Architecture and RNA Polymerization.
Nucleic Acids Res., 41, 2013
|
|
5MSF
 
 | | MS2 PROTEIN CAPSID/RNA COMPLEX | | Descriptor: | 5'-R(*CP*CP*GP*GP*AP*GP*GP*AP*UP*CP*AP*CP*CP*AP*CP*GP*GP*G)-3', MS2 PROTEIN CAPSID | | Authors: | Rowsell, S, Stonehouse, N.J, Convery, M.A, Adams, C.J, Ellington, A.D, Hirao, I, Peabody, D.S, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 1998-05-15 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of a series of RNA aptamers complexed to the same protein target.
Nat.Struct.Biol., 5, 1998
|
|
8C6D
 
 | | Production of antigenically stable enterovirus A71 virus-like particles in Pichia pastoris as a vaccine candidate. | | Descriptor: | (2S,3R,4E)-2-aminooctadec-4-ene-1,3-diol, Genome polyprotein, Genome polyprotein (Fragment) | | Authors: | Kingston, N.J, Snowden, J.S, Stonehouse, N.J, Rowlands, D.J, Hogle, J.M. | | Deposit date: | 2023-01-11 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Production of antigenically stable enterovirus A71 virus-like particles in Pichia pastoris as a vaccine candidate.
Biorxiv, 2023
|
|
6MSF
 
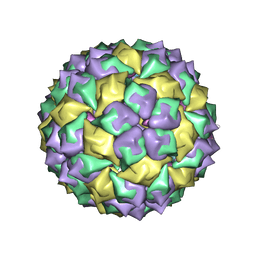 | | F6 APTAMER MS2 COAT PROTEIN COMPLEX | | Descriptor: | PROTEIN (MS2 PROTEIN CAPSID), RNA (5'-R(*CP*AP*GP*UP*CP*AP*CP*UP*GP*G)-3'), RNA (5'-R(*CP*CP*AP*CP*AP*GP*UP*CP*AP*CP*UP*GP*GP*G)-3') | | Authors: | Convery, M.A, Rowsell, S, Stonehouse, N.J, Ellington, A.D, Hirao, I, Murray, J.B, Peabody, D.S, Phillips, S.E.V, Stockley, P.G. | | Deposit date: | 1998-01-06 | | Release date: | 1998-07-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of an RNA aptamer-protein complex at 2.8 A resolution.
Nat.Struct.Biol., 5, 1998
|
|
7ZQ8
 
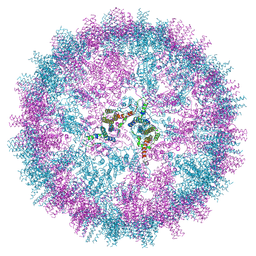 | | VelcroVax tandem HBcAg with SUMO-Affimer inserted at MIR (T=4 VLP) | | Descriptor: | VelcroVax tandem HBcAg with SUMO-Affimer inserted at MIR | | Authors: | Kingston, N.J, Grehan, K, Snowden, J.S, Alzahrani, J, Ranson, N.A, Rowlands, D.J, Stonehouse, N.J. | | Deposit date: | 2022-04-29 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | VelcroVax: a "Bolt-On" Vaccine Platform for Glycoprotein Display.
Msphere, 8, 2023
|
|
7BC4
 
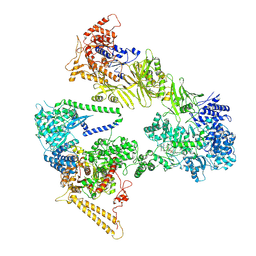 | | Cryo-EM structure of fatty acid synthase (FAS) from Pichia pastoris | | Descriptor: | FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta | | Authors: | Snowden, J.S, Alzahrani, J, Sherry, L, Stacey, M, Rowlands, D.J, Ranson, N.A, Stonehouse, N.J. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into Pichia pastoris fatty acid synthase.
Sci Rep, 11, 2021
|
|
7BC5
 
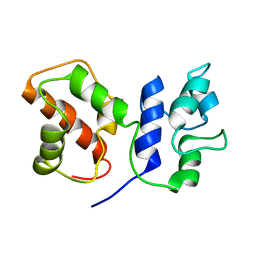 | | Cryo-EM structure of ACP domain from Pichia pastoris fatty acid synthase (FAS) | | Descriptor: | Fatty acid synthase subunit alpha | | Authors: | Snowden, J.S, Alzahrani, J, Sherry, L, Stacey, M, Rowlands, D.J, Ranson, N.A, Stonehouse, N.J. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into Pichia pastoris fatty acid synthase.
Sci Rep, 11, 2021
|
|
