6UVW
 
 | |
6UWK
 
 | |
6UWJ
 
 | |
6UWH
 
 | |
6UWG
 
 | |
6UW0
 
 | |
6CZG
 
 | |
6CZJ
 
 | |
3E54
 
 | | Archaeal Intron-encoded Homing Endonuclease I-Vdi141I Complexed With DNA | | Descriptor: | DNA (5'-D(*DCP*DTP*DGP*DAP*DCP*DTP*DCP*DTP*DCP*DTP*DTP*DAP*DA)-3'), DNA (5'-D(*DTP*DTP*DGP*DGP*DCP*DTP*DAP*DCP*DCP*DTP*DTP*DAP*DA)-3'), DNA (5'-D(P*DGP*DAP*DGP*DAP*DGP*DTP*DCP*DAP*DG)-3'), ... | | Authors: | Nomura, N, Nomura, Y, Sussman, D, Stoddard, B.L. | | Deposit date: | 2008-08-13 | | Release date: | 2008-12-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Recognition of a common rDNA target site in archaea and eukarya by analogous LAGLIDADG and His-Cys box homing endonucleases
Nucleic Acids Res., 36, 2008
|
|
6CZI
 
 | | Structure of a redesigned beta barrel, mFAP1, bound to DFHBI | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, mFAP1 | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2018-04-09 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
6CZH
 
 | | Structure of a redesigned beta barrel, mFAP0, bound to DFHBI | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, mFAP0 | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2018-04-09 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
6D0T
 
 | | De novo design of a fluorescence-activating beta barrel - BB1 | | Descriptor: | BB1 | | Authors: | Dou, J, Vorobieva, A.A, Sheffler, W, Doyle, L.A, Park, H, Bick, M.J, Mao, B, Foight, G.W, Lee, M, Carter, L, Sankaran, B, Ovchinnikov, S, Marcos, E, Huang, P, Vaughan, J.C, Stoddard, B.L, Baker, D. | | Deposit date: | 2018-04-10 | | Release date: | 2018-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
3E0L
 
 | | Computationally Designed Ammelide Deaminase | | Descriptor: | Guanine deaminase, ZINC ION | | Authors: | Murphy, P.M, Bolduc, J.M, Gallaher, J.L, Stoddard, B.L, Baker, D. | | Deposit date: | 2008-07-31 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Alteration of enzyme specificity by computational loop remodeling and design.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5ESP
 
 | |
5BYO
 
 | |
3EH8
 
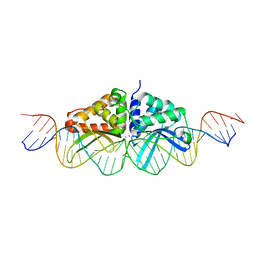 | |
4OUD
 
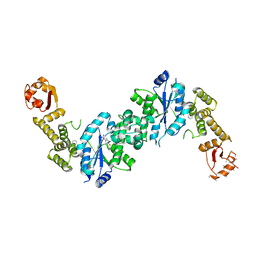 | | Engineered tyrosyl-tRNA synthetase with the nonstandard amino acid L-4,4-biphenylalanine | | Descriptor: | TYROSINE, Tyrosyl-tRNA synthetase | | Authors: | Takeuchi, R, Mandell, D.J, Lajoie, M.J, Church, G.M, Stoddard, B.L. | | Deposit date: | 2014-02-16 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Biocontainment of genetically modified organisms by synthetic protein design.
Nature, 518, 2015
|
|
1IDE
 
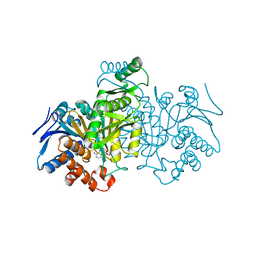 | | ISOCITRATE DEHYDROGENASE Y160F MUTANT STEADY-STATE INTERMEDIATE COMPLEX (LAUE DETERMINATION) | | Descriptor: | ISOCITRATE DEHYDROGENASE, ISOCITRIC ACID, MAGNESIUM ION, ... | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
1A73
 
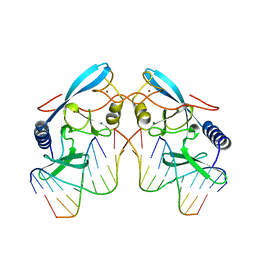 | | INTRON-ENCODED ENDONUCLEASE I-PPOI COMPLEXED WITH DNA | | Descriptor: | DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*TP*CP*TP*TP*AP*A)-3'), DNA (5'-D(P*GP*AP*GP*AP*GP*TP*CP*A)-3'), INTRON 3 (I-PPO) ENCODED ENDONUCLEASE, ... | | Authors: | Flick, K.E, Monnat Junior, R.J, Stoddard, B.L. | | Deposit date: | 1998-03-19 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI.
Nature, 394, 1998
|
|
4QPZ
 
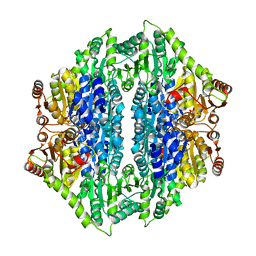 | | Crystal structure of the formolase FLS_v2 in space group P 21 | | Descriptor: | Formolase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Shen, B.W, Siegel, J.B, Stoddard, B.L, Baker, D. | | Deposit date: | 2014-06-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Computational protein design enables a novel one-carbon assimilation pathway.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4R6J
 
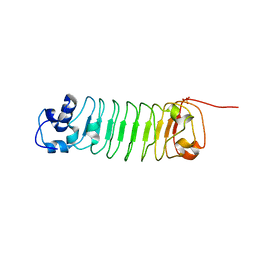 | |
4R6G
 
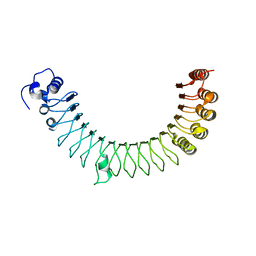 | |
8W0P
 
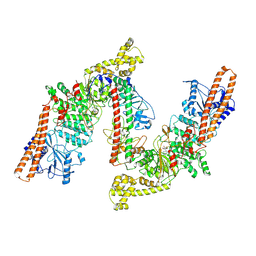 | | BsaXI -- Type IIB R-M system | | Descriptor: | RM.BsaXI, S-ADENOSYLMETHIONINE, S.BsaXI | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Atomic insights into the mechanism of DNA recognition and methylation by type IIB restriction-modification system: CryoEM structures of DNA free and cognate DNA bound BsaXI.
To Be Published
|
|
7SQ4
 
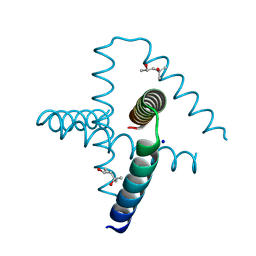 | | Designed trefoil knot protein, variant 2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Designed trefoil knot protein, variant 2, ... | | Authors: | Takushi, B, Doyle, L, Stoddard, B.L, Bradley, P. | | Deposit date: | 2021-11-04 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.493 Å) | | Cite: | De novo design of knotted tandem repeat proteins.
Nat Commun, 14, 2023
|
|
7SQ3
 
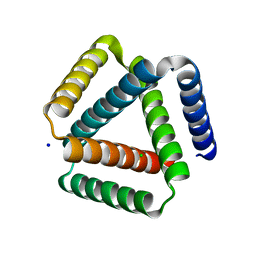 | | Designed trefoil knot protein, variant 1 | | Descriptor: | CHLORIDE ION, Designed trefoil knot protein, variant 1, ... | | Authors: | Takushi, B, Doyle, L, Stoddard, B.L, Bradley, P. | | Deposit date: | 2021-11-04 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.453 Å) | | Cite: | De novo design of knotted tandem repeat proteins.
Nat Commun, 14, 2023
|
|
