7B5G
 
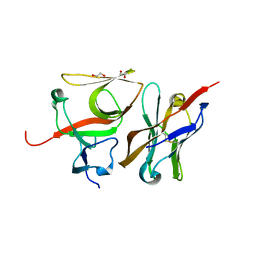 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS3(Nb14527) | | Descriptor: | 1,2-ETHANEDIOL, LexA repressor, Nanobody Nb14527, ... | | Authors: | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | Deposit date: | 2020-12-03 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
2BU4
 
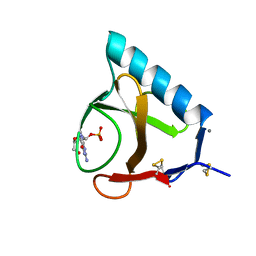 | | RIBONUCLEASE T1 COMPLEX WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Loris, R, Devos, S, Langhorst, U, Decanniere, K, Bouckaert, J, Maes, D, Transue, T.R, Steyaert, J. | | Deposit date: | 1998-09-14 | | Release date: | 1998-09-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Conserved water molecules in a large family of microbial ribonucleases.
Proteins, 36, 1999
|
|
7ZC2
 
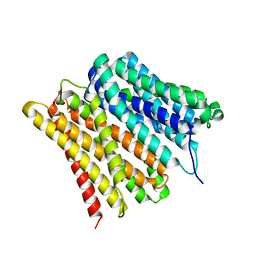 | | Dipeptide and tripeptide Permease C (DtpC) | | Descriptor: | Amino acid/peptide transporter | | Authors: | Killer, M, Finocchio, G, Pardon, E, Steyaert, J, Loew, C. | | Deposit date: | 2022-03-25 | | Release date: | 2022-07-06 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Cryo-EM Structure of an Atypical Proton-Coupled Peptide Transporter: Di- and Tripeptide Permease C.
Front Mol Biosci, 9, 2022
|
|
2BIR
 
 | |
5C1M
 
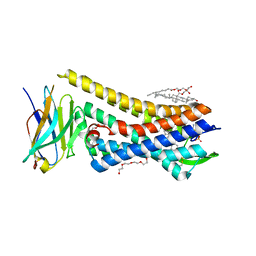 | | Crystal structure of active mu-opioid receptor bound to the agonist BU72 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R,3S,3aR,5aR,6R,11bR,11cS)-3a-methoxy-3,14-dimethyl-2-phenyl-2,3,3a,6,7,11c-hexahydro-1H-6,11b-(epiminoethano)-3,5a-methanonaphtho[2,1-g]indol-10-ol, CHOLESTEROL, ... | | Authors: | Huang, W.J, Manglik, A, Venkatakrishnan, A.J, Laeremans, T, Feinberg, E.N, Sanborn, A.L, Kato, H.E, Livingston, K.E, Thorsen, T.S, Kling, R, Granier, S, Gmeiner, P, Husbands, S.M, Traynor, J.R, Weis, W.I, Steyaert, J, Dror, R.O, Kobilka, B.K. | | Deposit date: | 2015-06-15 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural insights into mu-opioid receptor activation.
Nature, 524, 2015
|
|
6B73
 
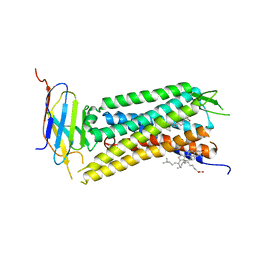 | | Crystal Structure of a nanobody-stabilized active state of the kappa-opioid receptor | | Descriptor: | CHOLESTEROL, N-[(5alpha,6beta)-17-(cyclopropylmethyl)-3-hydroxy-7,8-didehydro-4,5-epoxymorphinan-6-yl]-3-iodobenzamide, Nanobody, ... | | Authors: | Che, T, Majumdar, S, Zaidi, S.A, Kormos, C, McCorvy, J.D, Wang, S, Mosier, P.D, Uprety, R, Vardy, E, Krumm, B.E, Han, G.W, Lee, M.Y, Pardon, E, Steyaert, J, Huang, X.P, Strachan, R.T, Tribo, A.R, Pasternak, G.W, Carroll, I.F, Stevens, R.C, Cherezov, V, Katritch, V, Wacker, D, Roth, B.L. | | Deposit date: | 2017-10-03 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Nanobody-Stabilized Active State of the Kappa Opioid Receptor.
Cell, 172, 2018
|
|
2NXW
 
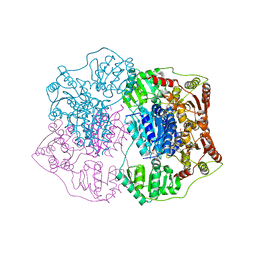 | | Crystal structure of phenylpyruvate decarboxylase of Azospirillum brasilense | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Versees, W, Spaepen, S, Vanderleyden, J, Steyaert, J. | | Deposit date: | 2006-11-20 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of phenylpyruvate decarboxylase from Azospirillum brasilense at 1.5 A resolution. Implications for its catalytic and regulatory mechanism.
Febs J., 274, 2007
|
|
6C9W
 
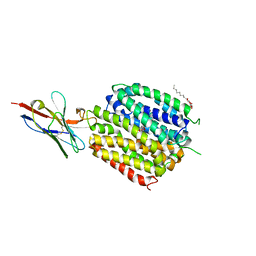 | | Crystal Structure of a ligand bound LacY/Nanobody Complex | | Descriptor: | 4-nitrophenyl alpha-D-galactopyranoside, Lactose permease, Nanobody9047, ... | | Authors: | Kumar, H, Finer-Moore, J.S, Jiang, X, Smirnova, I, Kasho, V, Pardon, E, Steyaert, J, Kaback, H.R, Stroud, R.M. | | Deposit date: | 2018-01-29 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of a ligand-bound LacY-Nanobody Complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6MXT
 
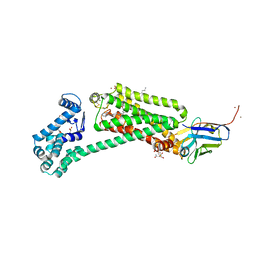 | | Crystal structure of human beta2 adrenergic receptor bound to salmeterol and Nb71 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Endolysin, ... | | Authors: | Masureel, M, Zou, Y, Picard, L.P, van der Westhuizen, E, Mahoney, J.P, Rodrigues, J.P.G.L.M, Mildorf, T.J, Dror, R.O, Shaw, D.E, Bouvier, M, Pardon, E, Steyaert, J, Sunahara, R.K, Weis, W.I, Zhang, C, Kobilka, B.K. | | Deposit date: | 2018-10-31 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95934224 Å) | | Cite: | Structural insights into binding specificity, efficacy and bias of a beta2AR partial agonist.
Nat. Chem. Biol., 14, 2018
|
|
3P0G
 
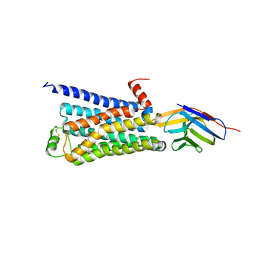 | | Structure of a nanobody-stabilized active state of the beta2 adrenoceptor | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Beta-2 adrenergic receptor, Lysozyme, ... | | Authors: | Rasmussen, S.G.F, Choi, H.-J, Fung, J.J, Pardon, E, Casarosa, P, Chae, P.S, DeVree, B.T, Rosenbaum, D.M, Thian, F.S, Kobilka, T.S, Schnapp, A, Konetzki, I, Sunahara, R.K, Gellman, S.H, Pautsch, A, Steyaert, J, Weis, W.I, Kobilka, B.K. | | Deposit date: | 2010-09-28 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a nanobody-stabilized active state of the b2 adrenoceptor
Nature, 469, 2011
|
|
3SN6
 
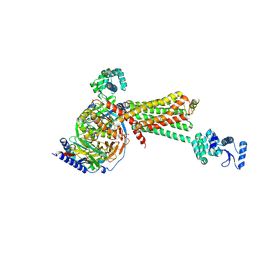 | | Crystal structure of the beta2 adrenergic receptor-Gs protein complex | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Camelid antibody VHH fragment, Endolysin,Beta-2 adrenergic receptor, ... | | Authors: | Rasmussen, S.G.F, DeVree, B.T, Zou, Y, Kruse, A.C, Chung, K.Y, Kobilka, T.S, Thian, F.S, Chae, P.S, Pardon, E, Calinski, D, Mathiesen, J.M, Shah, S.T.A, Lyons, J.A, Caffrey, M, Gellman, S.H, Steyaert, J, Skiniotis, G, Weis, W.I, Sunahara, R.K, Kobilka, B.K. | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the beta2 adrenergic receptor-Gs protein complex
Nature, 477, 2011
|
|
3ZBI
 
 | | Fitting result in the O-layer of the subnanometer structure of the bacterial pKM101 type IV secretion system core complex digested with elastase | | Descriptor: | TRAF PROTEIN, TRAN PROTEIN, TRAO PROTEIN | | Authors: | Rivera-Calzada, A, Fronzes, R, Savva, C.G, Chandran, V, Lian, P.W, Laeremans, T, Pardon, E, Steyaert, J, Remaut, H, Waksman, G, Orlova, E.V. | | Deposit date: | 2012-11-10 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Structure of a Bacterial Type Iv Secretion Core Complex at Subnanometre Resolution.
Embo J., 32, 2013
|
|
3ZBJ
 
 | | Fitting results in the I-layer of the subnanometer structure of the bacterial pKM101 type IV secretion system core complex digested with elastase | | Descriptor: | TRAO PROTEIN | | Authors: | Rivera-Calzada, A, Fronzes, R, Savva, C.G, Chandran, V, Lian, P.W, Laeremans, T, Pardon, E, Steyaert, J, Remaut, H, Waksman, G, Orlova, E.V. | | Deposit date: | 2012-11-10 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Structure of a Bacterial Type Iv Secretion Core Complex at Subnanometre Resolution.
Embo J., 32, 2013
|
|
6QD6
 
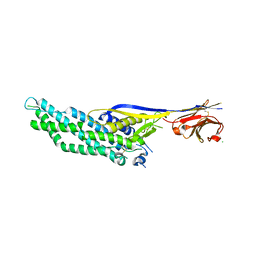 | | Molecular scaffolds expand the nanobody toolkit for cryo-EM applications: crystal structure of Mb-cHopQ-Nb207 | | Descriptor: | CHLORIDE ION, Mb-cHopQ-Nb207,Outer membrane protein,Mb-cHopQ-Nb207,Outer membrane protein,Mb-cHopQ-Nb207 | | Authors: | Uchanski, T, Masiulis, S, Fischer, B, Kalichuk, V, Wohlkonig, A, Zogg, T, Remaut, H, Vranken, W, Aricescu, A.R, Pardon, E, Steyaert, J. | | Deposit date: | 2018-12-31 | | Release date: | 2019-12-18 | | Last modified: | 2021-01-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM
Nat.Methods, 18, 2021
|
|
1RHL
 
 | | RIBONUCLEASE T1 COMPLEXED WITH 2'GMP/G23A MUTANT | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Huyghues-Despointes, B.M.P, Langhorst, U, Steyaert, J, Pace, C.N, Scholtz, J.M. | | Deposit date: | 1998-10-09 | | Release date: | 1998-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Hydrogen-exchange stabilities of RNase T1 and variants with buried and solvent-exposed Ala --> Gly mutations in the helix.
Biochemistry, 38, 1999
|
|
1R4F
 
 | | Inosine-Adenosine-Guanosine Preferring Nucleoside Hydrolase From Trypanosoma vivax: Trp260Ala Mutant In Complex With 3-Deaza-Adenosine | | Descriptor: | 3-DEAZA-ADENOSINE, CALCIUM ION, IAG-nucleoside hydrolase | | Authors: | Versees, W, Loverix, S, Vandemeulebroucke, A, Geerlings, P, Steyaert, J. | | Deposit date: | 2003-10-06 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Leaving group activation by aromatic stacking: an alternative to general Acid catalysis.
J.Mol.Biol., 338, 2004
|
|
5BOP
 
 | | Crystal structure of the artificial nanobody octarellinV.1 complex | | Descriptor: | Nanobody, Octarellin V.1 | | Authors: | Figueroa, M, Sleutel, M, Pardon, E, Steyaert, J, Martial, J.A, van de Weerdt, C. | | Deposit date: | 2015-05-27 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The unexpected structure of the designed protein Octarellin V.1 forms a challenge for protein structure prediction tools.
J.Struct.Biol., 195, 2016
|
|
5BIR
 
 | |
5BU4
 
 | | RIBONUCLEASE T1 COMPLEX WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Loris, R, Devos, S, Langhorst, U, Decanniere, K, Bouckaert, J, Maes, D, Transue, T.R, Steyaert, J. | | Deposit date: | 1998-09-15 | | Release date: | 1998-09-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Conserved water molecules in a large family of microbial ribonucleases.
Proteins, 36, 1999
|
|
6QFA
 
 | | CryoEM structure of a beta3K279T GABA(A)R homomer in complex with histamine and megabody Mb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit beta-3,Gamma-aminobutyric acid receptor subunit beta-3, HISTAMINE, ... | | Authors: | Uchanski, T, Masiulis, S, Fischer, B, Kalichuk, V, Wohlkoening, A, Zoegg, T, Remaut, H, Vranken, W, Aricescu, A.R, Pardon, E, Steyaert, J. | | Deposit date: | 2019-01-09 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM.
Nat.Methods, 18, 2021
|
|
2FF2
 
 | | Crystal structure of Trypanosoma vivax nucleoside hydrolase co-crystallized with ImmucillinH | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, CALCIUM ION, IAG-nucleoside hydrolase, ... | | Authors: | Versees, W, Barlow, J, Steyaert, J. | | Deposit date: | 2005-12-18 | | Release date: | 2006-05-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Transition-state Complex of the Purine-specific Nucleoside Hydrolase of T.vivax: Enzyme Conformational Changes and Implications for Catalysis.
J.Mol.Biol., 359, 2006
|
|
1BU4
 
 | | RIBONUCLEASE 1 COMPLEX WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Loris, R, Devos, S, Langhorst, U, Decanniere, K, Bouckaert, J, Maes, D, Transue, T.R, Steyaert, J. | | Deposit date: | 1998-09-11 | | Release date: | 1999-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conserved water molecules in a large family of microbial ribonucleases.
Proteins, 36, 1999
|
|
1BVI
 
 | | RIBONUCLEASE T1 (WILDTYPE) COMPLEXED WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Langhorst, U, Loris, R, Denisov, V.P, Doumen, J, Roose, P, Maes, D, Halle, B, Steyaert, J. | | Deposit date: | 1998-09-15 | | Release date: | 1998-09-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dissection of the structural and functional role of a conserved hydration site in RNase T1.
Protein Sci., 8, 1999
|
|
1FYS
 
 | | Ribonuclease T1 V16C mutant | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, GUANYL-SPECIFIC RIBONUCLEASE T1 | | Authors: | De Vos, S, Loris, R, Steyaert, J. | | Deposit date: | 2000-10-03 | | Release date: | 2000-10-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hydrophobic core manipulations in ribonuclease T1.
Biochemistry, 40, 2001
|
|
1FZU
 
 | | RNAse T1 V78A mutant | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, GUANYL-SPECIFIC RIBONUCLEASE T1 | | Authors: | De Vos, S, Loris, R, Steyaert, J. | | Deposit date: | 2000-10-04 | | Release date: | 2000-10-25 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hydrophobic core manipulations in ribonuclease T1.
Biochemistry, 40, 2001
|
|
