1LTU
 
 | | CRYSTAL STRUCTURE OF CHROMOBACTERIUM VIOLACEUM, APO (NO IRON BOUND) STRUCTURE | | Descriptor: | PHENYLALANINE-4-HYDROXYLASE | | Authors: | Erlandsen, H, Kim, J.Y, Patch, M.G, Han, A, Volner, A, Abu-Omar, M.M, Stevens, R.C. | | Deposit date: | 2002-05-20 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates.
J.Mol.Biol., 320, 2002
|
|
1LTZ
 
 | | CRYSTAL STRUCTURE OF CHROMOBACTERIUM VIOLACEUM PHENYLALANINE HYDROXYLASE, STRUCTURE HAS BOUND IRON (III) AND OXIDIZED COFACTOR 7,8-DIHYDROBIOPTERIN | | Descriptor: | 7,8-DIHYDROBIOPTERIN, CHLORIDE ION, FE (III) ION, ... | | Authors: | Erlandsen, H, Kim, J.Y, Patch, M.G, Han, A, Volner, A, Abu-Omar, M.M, Stevens, R.C. | | Deposit date: | 2002-05-21 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates.
J.Mol.Biol., 320, 2002
|
|
2WAP
 
 | | 3D-crystal structure of humanized-rat fatty acid amide hydrolase (FAAH) conjugated with the drug-like urea inhibitor PF-3845 | | Descriptor: | 4-(3-{[5-(trifluoromethyl)pyridin-2-yl]oxy}benzyl)piperidine-1-carboxylic acid, CHLORIDE ION, FATTY-ACID AMIDE HYDROLASE 1, ... | | Authors: | Mileni, M, Kamtekar, S, Stevens, R.C. | | Deposit date: | 2009-02-11 | | Release date: | 2009-05-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Characterization of a Highly Selective Faah Inhibitor that Reduces Inflammatory Pain.
Chem.Biol., 16, 2009
|
|
2VI6
 
 | | Crystal Structure of the Nanog Homeodomain | | Descriptor: | HOMEOBOX PROTEIN NANOG | | Authors: | Jauch, R, Ng, C.K.L, Saitakendu, K.S, Stevens, R.C, Kolatkar, P.R. | | Deposit date: | 2007-11-28 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure and DNA Binding of the Homeodomain of the Stem Cell Transcription Factor Nanog.
J.Mol.Biol., 376, 2008
|
|
2VU9
 
 | | CRYSTAL STRUCTURE OF BOTULINUM NEUROTOXIN SEROTYPE A BINDING DOMAIN IN COMPLEX WITH GT1B | | Descriptor: | BOTULINUM NEUROTOXIN A HEAVY CHAIN, MAGNESIUM ION, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Stenmark, P, Dupuy, J, Stevens, R.C. | | Deposit date: | 2008-05-22 | | Release date: | 2008-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Botulinum Neurotoxin Type a in Complex with the Cell Surface Co-Receptor Gt1B- Insight Into the Toxin-Neuron Interaction.
Plos Pathog., 4, 2008
|
|
1LTV
 
 | | CRYSTAL STRUCTURE OF CHROMOBACTERIUM VIOLACEUM PHENYLALANINE HYDROXYLASE, STRUCTURE WITH BOUND OXIDIZED Fe(III) | | Descriptor: | FE (III) ION, PHENYLALANINE-4-HYDROXYLASE | | Authors: | Erlandsen, H, Kim, J.Y, Patch, M.G, Han, A, Volner, A, Abu-Omar, M.M, Stevens, R.C. | | Deposit date: | 2002-05-20 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates.
J.Mol.Biol., 320, 2002
|
|
2N2F
 
 | | Solution NMR structure of Dynorphin 1-13 bound to Kappa Opioid Receptor | | Descriptor: | Dynorphin A(1-13) | | Authors: | O'Connor, C, White, K, Doncescu, N, Didenko, T, Roth, B.L, Czaplicki, G, Stevens, R.C, Wuthrich, K, Milon, A. | | Deposit date: | 2015-05-06 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of the agonist dynorphin peptide bound to the human kappa opioid receptor.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1MT5
 
 | | CRYSTAL STRUCTURE OF FATTY ACID AMIDE HYDROLASE | | Descriptor: | Fatty-acid amide hydrolase, METHYL ARACHIDONYL FLUOROPHOSPHONATE | | Authors: | Bracey, M.H, Hanson, M.A, Masuda, K.R, Stevens, R.C, Cravatt, B.F. | | Deposit date: | 2002-09-20 | | Release date: | 2002-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Adaptations in a Membrane Enzyme That Terminates Endocannabinoid Signaling
science, 298, 2002
|
|
2FPQ
 
 | | Crystal Structure of Botulinum Neurotoxin Type D Light Chain | | Descriptor: | BOTULINUM NEUROTOXIN D LIGHT CHAIN, POTASSIUM ION, ZINC ION | | Authors: | Arndt, J.W, Chai, Q, Christian, T, Stevens, R.C. | | Deposit date: | 2006-01-16 | | Release date: | 2006-03-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Botulinum Neurotoxin Type D Light Chain at 1.65 A Resolution: Repercussions for VAMP-2 Substrate Specificity(,).
Biochemistry, 45, 2006
|
|
2FYG
 
 | | Crystal structure of NSP10 from Sars coronavirus | | Descriptor: | GLYCEROL, Replicase polyprotein 1ab, ZINC ION | | Authors: | Joseph, J.S, Saikatendu, K.S, Subramanian, V, Neuman, B.W, Brooun, A, Griffith, M, Moy, K, Yadav, M.K, Velasquez, J, Buchmeier, M.J, Stevens, R.C, Kuhn, P. | | Deposit date: | 2006-02-07 | | Release date: | 2006-08-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs.
J.Virol., 80, 2006
|
|
1TG2
 
 | | Crystal structure of phenylalanine hydroxylase A313T mutant with 7,8-dihydrobiopterin bound | | Descriptor: | 2-AMINO-6-(1,2-DIHYDROXY-PROPYL)-7,8-DIHYDRO-6H-PTERIDIN-4-ONE, FE (III) ION, Phenylalanine-4-hydroxylase | | Authors: | Erlandsen, H, Pey, A.L, Gamez, A, Perez, B, Desviat, L.R, Aguado, C, Koch, R, Surendran, S, Tyring, S, Matalon, R, Scriver, C.R, Ugarte, M, Martinez, A, Stevens, R.C. | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Correction of kinetic and stability defects by tetrahydrobiopterin in phenylketonuria patients with certain phenylalanine hydroxylase mutations.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1TDW
 
 | | Crystal structure of double truncated human phenylalanine hydroxylase BH4-responsive PKU mutant A313T. | | Descriptor: | FE (III) ION, Phenylalanine-4-hydroxylase | | Authors: | Erlandsen, H, Pey, A.L, Gamez, A, Perez, B, Desviat, L.R, Aguado, C, Koch, R, Surendran, S, Tyring, S, Matalon, R, Scriver, C.R, Ugarte, M, Martinez, A, Stevens, R.C. | | Deposit date: | 2004-05-24 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Correction of kinetic and stability defects by tetrahydrobiopterin in phenylketonuria patients with certain phenylalanine hydroxylase mutations.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
2TOH
 
 | | TYROSINE HYDROXYLASE CATALYTIC AND TETRAMERIZATION DOMAINS FROM RAT | | Descriptor: | 7,8-DIHYDROBIOPTERIN, CHLORIDE ION, FE (III) ION, ... | | Authors: | Goodwill, K.E, Sabatier, C, Stevens, R.C. | | Deposit date: | 1998-08-26 | | Release date: | 1999-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of tyrosine hydroxylase with bound cofactor analogue and iron at 2.3 A resolution: self-hydroxylation of Phe300 and the pterin-binding site.
Biochemistry, 37, 1998
|
|
1D5B
 
 | | UNLIGANDED MATURE OXY-COPE CATALYTIC ANTIBODY | | Descriptor: | CADMIUM ION, chimeric OXY-COPE catalytic ANTIBODY AZ-28 (HEAVY chain), chimeric OXY-COPE catalytic ANTIBODY AZ-28 (light chain) | | Authors: | Mundorff, E.C, Hanson, M.A, Schultz, P.G, Stevens, R.C. | | Deposit date: | 1999-10-06 | | Release date: | 2000-02-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational effects in biological catalysis: an antibody-catalyzed oxy-cope rearrangement.
Biochemistry, 39, 2000
|
|
1GAF
 
 | | 48G7 HYBRIDOMA LINE FAB COMPLEXED WITH HAPTEN 5-(PARA-NITROPHENYL PHOSPHONATE)-PENTANOIC ACID | | Descriptor: | 5-(PARA-NITROPHENYL PHOSPHONATE)-PENTANOIC ACID, CHIMERIC 48G7 FAB | | Authors: | Wedemayer, G.J, Patten, P.A, Stevens, R.C, Schultz, P.G. | | Deposit date: | 1996-02-06 | | Release date: | 1996-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The immunological evolution of catalysis.
Science, 271, 1996
|
|
1F82
 
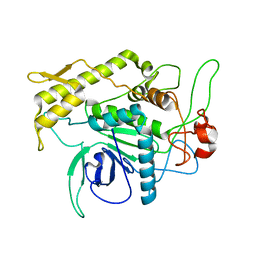 | | BOTULINUM NEUROTOXIN TYPE B CATALYTIC DOMAIN | | Descriptor: | BOTULINUM NEUROTOXIN TYPE B, ZINC ION | | Authors: | Hanson, M.A, Stevens, R.C. | | Deposit date: | 2000-06-28 | | Release date: | 2000-08-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cocrystal structure of synaptobrevin-II bound to botulinum neurotoxin type B at 2.0 A resolution.
Nat.Struct.Biol., 7, 2000
|
|
2WJ2
 
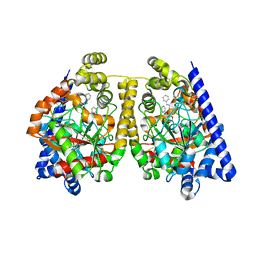 | | 3D-crystal structure of humanized-rat fatty acid amide hydrolase (FAAH) conjugated with 7-phenyl-1-(5-(pyridin-2-yl)oxazol-2-yl)heptan- 1-one, an alpha-ketooxazole | | Descriptor: | 7-phenyl-1-(5-pyridin-2-yl-1,3-oxazol-2-yl)heptane-1,1-diol, CHLORIDE ION, FATTY ACID AMIDE HYDROLASE 1 | | Authors: | Mileni, M, Garfunkle, J, DeMartino, J.K, Cravatt, B.F, Boger, D.L, Stevens, R.C. | | Deposit date: | 2009-05-19 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Binding and Inactivation Mechanism of a Humanized Fatty Acid Amide Hydrolase by Alpha-Ketoheterocycle Inhibitors Revealed from Cocrystal Structures.
J.Am.Chem.Soc., 131, 2009
|
|
2WJ1
 
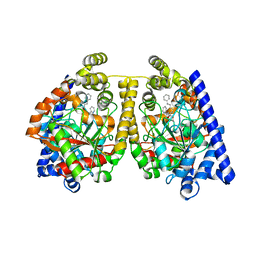 | | 3D-crystal structure of humanized-rat fatty acid amide hydrolase (FAAH) conjugated with 7-phenyl-1-(4-(pyridin-2-yl)oxazol-2-yl)heptan- 1-one, an alpha-ketooxazole | | Descriptor: | 7-phenyl-1-(4-pyridin-2-yl-1,3-oxazol-2-yl)heptane-1,1-diol, CHLORIDE ION, FATTY-ACID AMIDE HYDROLASE 1 | | Authors: | Mileni, M, Garfunkle, J, DeMartino, J.K, Cravatt, B.F, Boger, D.L, Stevens, R.C. | | Deposit date: | 2009-05-19 | | Release date: | 2009-09-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Binding and Inactivation Mechanism of a Humanized Fatty Acid Amide Hydrolase by Alpha-Ketoheterocycle Inhibitors Revealed from Cocrystal Structures.
J.Am.Chem.Soc., 131, 2009
|
|
1D5I
 
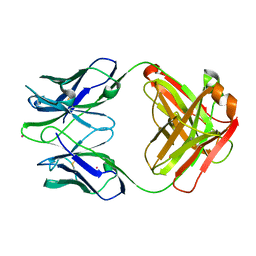 | | UNLIGANDED GERMLINE PRECURSOR OF AN OXY-COPE CATALYTIC ANTIBODY | | Descriptor: | CADMIUM ION, CHIMERIC GERMLINE PRECURSOR OF OXY-COPE CATALYTIC ANTIBODY AZ-28 (HEAVY CHAIN), CHIMERIC GERMLINE PRECURSOR OF OXY-COPE CATALYTIC ANTIBODY AZ-28 (LIGHT CHAIN) | | Authors: | Mundorff, E.C, Hanson, M.A, Schultz, P.G, Stevens, R.C. | | Deposit date: | 1999-10-07 | | Release date: | 2000-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational effects in biological catalysis: an antibody-catalyzed oxy-cope rearrangement.
Biochemistry, 39, 2000
|
|
1N7M
 
 | | Germline 7G12 with N-methylmesoporphyrin | | Descriptor: | Germline Metal Chelatase Catalytic Antibody, chain H, chain L, ... | | Authors: | Yin, J, Andryski, S.E, Beuscher IV, A.E, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2002-11-15 | | Release date: | 2003-02-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evidence for substrate strain in antibody catalysis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NGX
 
 | | Chimeric Germline Fab 7g12 with jeffamine fragment bound | | Descriptor: | Germline Metal Chelatase Catalytic Antibody, Heavy chain, Light chain, ... | | Authors: | Yin, J, Andryski, S.E, Beuscher, A.B, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2002-12-18 | | Release date: | 2003-03-18 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evidence for substrate strain in antibody catalysis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1TOH
 
 | |
1UB5
 
 | | Crystal structure of Antibody 19G2 with hapten at 100K | | Descriptor: | 4-(4-STYRYL-PHENYLCARBAMOYL)-BUTYRIC ACID, antibody 19G2, alpha chain, ... | | Authors: | Beuscher, A.B, Wirsching, P, Lerner, R.A, Janda, K, Stevens, R.C. | | Deposit date: | 2003-03-30 | | Release date: | 2004-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Dynamics of Blue Fluorescent Antibody 19G2 at Blue and Violet Fluorescent Temperatures
To be published
|
|
2GRI
 
 | | NMR Structure of the SARS-CoV non-structural protein nsp3a | | Descriptor: | NSP3 | | Authors: | Serrano, P, Almeida, M.S, Johnson, M.A, Herrmann, T, Saikatendu, K.S, Joseph, J, Subramanian, V, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-04-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
2IDY
 
 | | NMR Structure of the SARS-CoV non-structural protein nsp3a | | Descriptor: | NSP3 | | Authors: | Serrano, P, Almeida, M.S, Johnson, M.A, Horst, R, Herrmann, T, Joseph, J, Saikatendu, K, Subramanian, V, Stevens, R.C, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-09-15 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
