3OVA
 
 | | How the CCA-adding Enzyme Selects Adenine over Cytosine in Position 76 of tRNA | | Descriptor: | 1,2-ETHANEDIOL, CCA-adding enzyme, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Pan, B.C, Xiong, Y, Steitz, T.A. | | Deposit date: | 2010-09-16 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | How the CCA-Adding Enzyme Selects Adenine over Cytosine at Position 76 of tRNA.
Science, 330, 2010
|
|
4WQY
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with elongation factor G in the post-translocational state (without fusitic acid) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Lin, J, Gagnon, M.G, Steitz, T.A. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational Changes of Elongation Factor G on the Ribosome during tRNA Translocation.
Cell, 160, 2015
|
|
4WPO
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with elongation factor G in the pre-translocational state | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Lin, J, Gagnon, M.G, Steitz, T.A. | | Deposit date: | 2014-10-20 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational Changes of Elongation Factor G on the Ribosome during tRNA Translocation.
Cell, 160, 2015
|
|
1DPI
 
 | |
4WQU
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with elongation factor G trapped by the antibiotic dityromycin | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Lin, J, Gagnon, M.G, Steitz, T.A. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational Changes of Elongation Factor G on the Ribosome during tRNA Translocation.
Cell, 160, 2015
|
|
4WQF
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with elongation factor G and fusidic acid in the post-translocational state | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Lin, J, Gagnon, M.G, Steitz, T.A. | | Deposit date: | 2014-10-21 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational Changes of Elongation Factor G on the Ribosome during tRNA Translocation.
Cell, 160, 2015
|
|
4Y4O
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome with rRNA modifications and bound to protein Y (YfiA) at 2.3A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Polikanov, Y.S, Melnikov, S.V, Soll, D, Steitz, T.A. | | Deposit date: | 2015-02-10 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4Y4P
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome with rRNA modifications and bound to mRNA and A-, P- and E-site tRNAs at 2.5A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Melnikov, S.V, Soll, D, Steitz, T.A. | | Deposit date: | 2015-02-10 | | Release date: | 2015-03-18 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly.
Nat.Struct.Mol.Biol., 22, 2015
|
|
1HKG
 
 | |
4Z8C
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome bound to translation inhibitor oncocin | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Roy, R.N, Lomakin, I.B, Gagnon, M.G, Steitz, T.A. | | Deposit date: | 2015-04-08 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The mechanism of inhibition of protein synthesis by the proline-rich peptide oncocin.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3OUY
 
 | | How the CCA-adding Enzyme Selects Adenine Over Cytosine at Position 76 of tRNA | | Descriptor: | 1,2-ETHANEDIOL, CCA-Adding Enzyme, PYROPHOSPHATE 2-, ... | | Authors: | Pan, B.C, Xiong, Y, Steitz, T.A. | | Deposit date: | 2010-09-15 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | How the CCA-Adding Enzyme Selects Adenine over Cytosine at Position 76 of tRNA.
Science, 330, 2010
|
|
3OV7
 
 | | How the CCA-Adding Enzyme Selects Adenine over Cytosine in Position 76 of tRNA | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CCA-Adding Enzyme, ... | | Authors: | Pan, B.C, Xiong, Y, Steitz, T.A. | | Deposit date: | 2010-09-15 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How the CCA-Adding Enzyme Selects Adenine over Cytosine at Position 76 of tRNA.
Science, 330, 2010
|
|
4Z3S
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with antibiotic A201A, mRNA and three tRNAs in the A, P and E sites at 2.65A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 3'-deoxy-3'-{[(2E)-3-(4-{[(4Z)-6-O-(6-deoxy-3,4-di-O-methyl-alpha-D-mannopyranosyl)-5-O-methyl-alpha-D-threo-hex-4-enofuranosyl]oxy}phenyl)-2-methylprop-2-enoyl]amino}-N,N-dimethyladenosine, ... | | Authors: | Polikanov, Y.S, Starosta, A.L, Juette, M.F, Altman, R.B, Terry, D.S, Lu, W, Burnett, B.J, Dinos, G, Reynolds, K, Blanchard, S.C, Steitz, T.A, Wilson, D.N. | | Deposit date: | 2015-03-31 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A.
Mol.Cell, 58, 2015
|
|
2EX3
 
 | | Bacteriophage phi29 DNA polymerase bound to terminal protein | | Descriptor: | DNA polymerase, DNA terminal protein, LEAD (II) ION | | Authors: | Kamtekar, S, Berman, A.J, Wang, J, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2005-11-07 | | Release date: | 2006-03-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The phi29 DNA polymerase:protein-primer structure suggests a model for the initiation to elongation transition
Embo J., 25, 2006
|
|
1XHX
 
 | | Phi29 DNA Polymerase, orthorhombic crystal form | | Descriptor: | DNA polymerase, MAGNESIUM ION, SULFATE ION | | Authors: | Kamtekar, S, Berman, A.J, Wang, J, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2004-09-21 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Insights into Strand Displacement and Processivity from the Crystal Structure of the Protein-Primed DNA Polymerase of Bacteriophage phi29
Mol.Cell, 16, 2004
|
|
2EFG
 
 | | TRANSLATIONAL ELONGATION FACTOR G COMPLEXED WITH GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, PROTEIN (ELONGATION FACTOR G DOMAIN 3), PROTEIN (ELONGATION FACTOR G) | | Authors: | Czworkowski, J, Wang, J, Steitz, T.A, Moore, P.B. | | Deposit date: | 1998-09-23 | | Release date: | 1999-09-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution.
EMBO J., 13, 1994
|
|
6B6H
 
 | | The cryo-EM structure of a bacterial class I transcription activation complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Hong, C, Huang, R, Yu, Z, Steitz, T.A. | | Deposit date: | 2017-10-02 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of bacterial transcription activation.
Science, 358, 2017
|
|
3G6E
 
 | | Co-crystal structure of Homoharringtonine bound to the large ribosomal subunit | | Descriptor: | (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, 23S ribosomal RNA, 50S ribosomal protein L10E, ... | | Authors: | Gurel, G, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2009-02-06 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome.
J.Mol.Biol., 389, 2009
|
|
1QRU
 
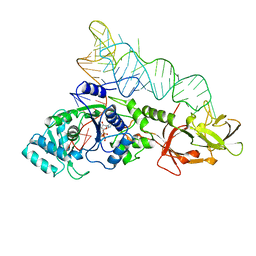 | |
3G71
 
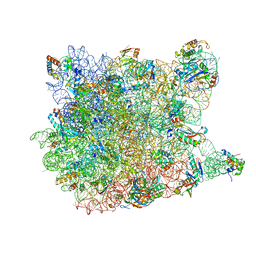 | | Co-crystal structure of Bruceantin bound to the large ribosomal subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10E, 50S ribosomal protein L10e, ... | | Authors: | Gurel, G, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2009-02-09 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome.
J.Mol.Biol., 389, 2009
|
|
1KLN
 
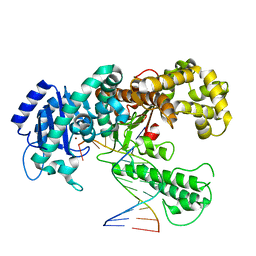 | | DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7) MUTANT/DNA COMPLEX | | Descriptor: | DNA (5'-D(*GP*CP*CP*GP*CP*GP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*CP*GP*CP*GP*GP*CP*GP*GP*C)-3'), PROTEIN (DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7)), ... | | Authors: | Beese, L.S, Derbyshire, V, Steitz, T.A. | | Deposit date: | 1994-05-24 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of DNA polymerase I Klenow fragment bound to duplex DNA.
Science, 260, 1993
|
|
2FOT
 
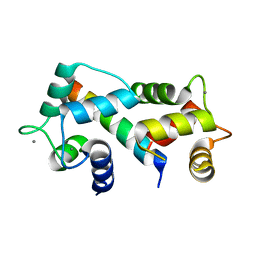 | | Crystal structure of the complex between calmodulin and alphaII-spectrin | | Descriptor: | CALCIUM ION, Calmodulin, alpha-II spectrin Spectrin | | Authors: | Simonovic, M, Zhang, Z, Cianci, C.D, Steitz, T.A, Morrow, J.S. | | Deposit date: | 2006-01-13 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the calmodulin alphaII-spectrin complex provides insight into the regulation of cell plasticity.
J.Biol.Chem., 281, 2006
|
|
3G4S
 
 | | Co-crystal structure of Tiamulin bound to the large ribosomal subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L10e, ... | | Authors: | Gurel, G, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2009-02-04 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome.
J.Mol.Biol., 389, 2009
|
|
1QRT
 
 | |
1QRS
 
 | |
