8CLG
 
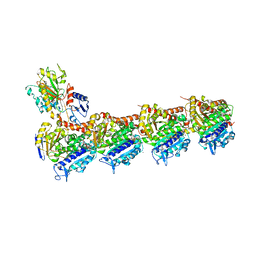 | | Epothilone A and Colchicine bound to tubulin (T2R-TTL) complex | | Descriptor: | CALCIUM ION, EPOTHILONE A, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wranik, M, Bertrand, Q, Kepa, M.W, Weinert, T, Steinmetz, M, Standfuss, J. | | Deposit date: | 2023-02-16 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A multi-reservoir extruder for time-resolved serial protein crystallography and compound screening at X-ray free-electron lasers.
Nat Commun, 14, 2023
|
|
8CLD
 
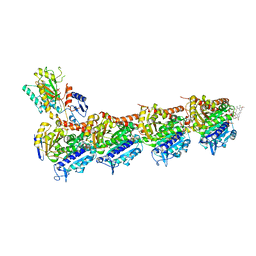 | | Ansamitocin P3 bound to tubulin (T2R-TTL) complex | | Descriptor: | (1S,2S,3S,5S,6S,16Z,18Z,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl 2-methylpropanoate, CALCIUM ION, Detyrosinated tubulin alpha-1B chain, ... | | Authors: | Wranik, M, Kepa, M.W, Bertrand, Q, Weinert, T, Steinmetz, M, Standfuss, J. | | Deposit date: | 2023-02-16 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A multi-reservoir extruder for time-resolved serial protein crystallography and compound screening at X-ray free-electron lasers.
Nat Commun, 14, 2023
|
|
8CLA
 
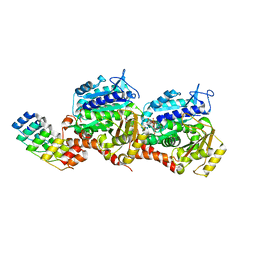 | | Z-SBTubA4 photoswitch bound to tubulin-DARPin D1 complex | | Descriptor: | 2-methoxy-5-[2-(5,6,7-trimethoxy-1,3-benzothiazol-2-yl)ethyl]phenol, CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Wranik, M, Bertrand, Q, Kepa, M.W, Weinert, T, Steinmetz, M, Standfuss, J. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A multi-reservoir extruder for time-resolved serial protein crystallography and compound screening at X-ray free electron lasers
To Be Published
|
|
6BR5
 
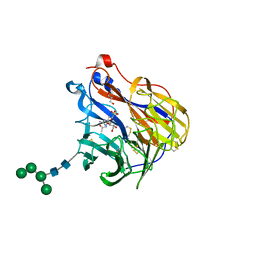 | | N2 neuraminidase in complex with a novel antiviral compound | | Descriptor: | (1R)-4-acetamido-1,5-anhydro-3-carbamimidamido-2,3,4-trideoxy-1-sulfo-D-glycero-D-galacto-octitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, L.H, Ve, T, Pascolutti, M, Hadhazi, A, Bailly, B, Thomson, R.J, Gao, G.F, von Itzstein, M. | | Deposit date: | 2017-11-30 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.0379076 Å) | | Cite: | A Sulfonozanamivir Analogue Has Potent Anti-influenza Virus Activity.
ChemMedChem, 13, 2018
|
|
6BR6
 
 | | N2 neuraminidase in complex with a novel antiviral compound | | Descriptor: | (1R)-4-acetamido-3-amino-1,5-anhydro-2,3,4-trideoxy-1-sulfo-D-glycero-D-galacto-octitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, L.H, Ve, T, Pascolutti, M, Hadhazi, A, Bailly, B, Thomson, R.J, Gao, G.F, von Itzstein, M. | | Deposit date: | 2017-11-30 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.03983879 Å) | | Cite: | A Sulfonozanamivir Analogue Has Potent Anti-influenza Virus Activity.
ChemMedChem, 13, 2018
|
|
6C0M
 
 | | The synthesis, biological evaluation and structural insights of unsaturated 3-N-substituted sialic acids as probes of human parainfluenza virus-3 haemagglutinin-neuraminidase | | Descriptor: | 1,2-ETHANEDIOL, 2,6-anhydro-3,5-dideoxy-5-[(2-methylpropanoyl)amino]-3-(4-phenyl-1H-1,2,3-triazol-1-yl)-D-glycero-D-galacto-non-2-enoni c acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dirr, L, Ve, T, von Itzstein, M. | | Deposit date: | 2018-01-01 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Insights into Human Parainfluenza Virus 3 Hemagglutinin-Neuraminidase Using Unsaturated 3- N-Substituted Sialic Acids as Probes.
ACS Chem. Biol., 13, 2018
|
|
1EIJ
 
 | | NMR ENSEMBLE OF METHANOBACTERIUM THERMOAUTOTROPHICUM PROTEIN 1615 | | Descriptor: | HYPOTHETICAL PROTEIN MTH1615 | | Authors: | Christendat, D, Booth, V, Gernstein, M, Arrowsmith, C.H, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-02-25 | | Release date: | 2000-11-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural proteomics of an archaeon.
Nat.Struct.Biol., 7, 2000
|
|
2UZ5
 
 | | Solution structure of the fkbp-domain of Legionella pneumophila Mip | | Descriptor: | MACROPHAGE INFECTIVITY POTENTIATOR | | Authors: | Ceymann, A, Horstmann, M, Ehses, P, Schweimer, K, Steinert, M, Kamphausen, T, Fischer, G, Hacker, J, Rosch, P, Faber, C. | | Deposit date: | 2007-04-25 | | Release date: | 2007-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Domain Motions of the Mip Protein from Legionella Pneumophila
Biochemistry, 45, 2006
|
|
4FBL
 
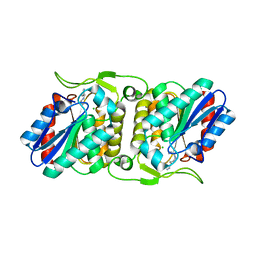 | | LipS and LipT, two metagenome-derived lipolytic enzymes increase the diversity of known lipase and esterase families | | Descriptor: | CHLORIDE ION, LipS lipolytic enzyme, SPERMIDINE | | Authors: | Chow, J, Krauss, U, Dall Antonia, Y, Fersini, F, Schmeisser, C, Schmidt, M, Menyes, I, Bornscheuer, U, Lauinger, B, Bongen, P, Pietruszka, J, Eckstein, M, Thum, O, Liese, A, Mueller-Dieckmann, J, Jaeger, K.-E, Kovacic, F, Streit, W.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Metagenome-Derived Enzymes LipS and LipT Increase the Diversity of Known Lipases.
Plos One, 7, 2012
|
|
6FUY
 
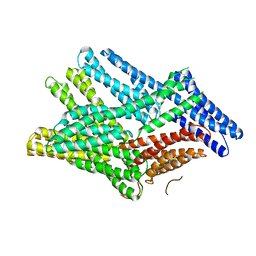 | | Crystal structure of human full-length vinculin-T12-A974K (residues 1-1066) | | Descriptor: | CALCIUM ION, Vinculin | | Authors: | Chorev, D.S, Volberg, T, Livne, A, Eisenstein, M, Martins, B, Kam, Z, Jockusch, B.M, Medalia, O, Sharon, M, Geiger, B. | | Deposit date: | 2018-02-28 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Conformational states during vinculin unlocking differentially regulate focal adhesion properties.
Sci Rep, 8, 2018
|
|
4FBM
 
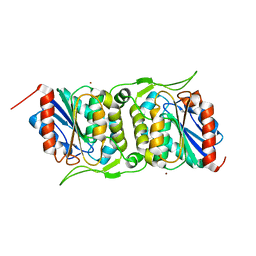 | | LipS and LipT, two metagenome-derived lipolytic enzymes increase the diversity of known lipase and esterase families | | Descriptor: | BROMIDE ION, LipS lipolytic enzyme | | Authors: | Chow, J, Krauss, U, Dall Antonia, Y, Fersini, F, Schmeisser, C, Schmidt, M, Menyes, I, Bornscheuer, U, Lauinger, B, Bongen, P, Pietruszka, J, Eckstein, M, Thum, O, Liese, A, Mueller-Dieckmann, J, Jaeger, K.-E, Kovavic, F, Streit, W.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Metagenome-Derived Enzymes LipS and LipT Increase the Diversity of Known Lipases.
Plos One, 7, 2012
|
|
1F8B
 
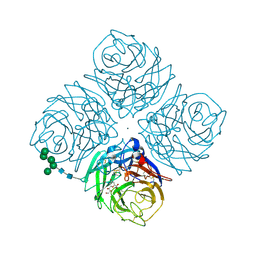 | | Native Influenza Virus Neuraminidase in Complex with NEU5AC2EN | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Smith, B.J, Colman, P.M, Von Itzstein, M, Danylec, B, Varghese, J.N. | | Deposit date: | 2000-06-30 | | Release date: | 2001-04-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of inhibitor binding in influenza virus neuraminidase.
Protein Sci., 10, 2001
|
|
1F8C
 
 | | Native Influenza Neuraminidase in Complex with 4-amino-2-deoxy-2,3-dehydro-N-neuraminic Acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-AMINO-2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, ... | | Authors: | Smith, B.J, Colman, P.M, Von Itzstein, M, Danylec, B, Varghese, J.N. | | Deposit date: | 2000-06-30 | | Release date: | 2001-04-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analysis of inhibitor binding in influenza virus neuraminidase.
Protein Sci., 10, 2001
|
|
1F8D
 
 | | Native Influenza Neuraminidase in Complex with 9-amino-2-deoxy-2,3-dehydro-N-neuraminic Acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-AMINO-2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, ... | | Authors: | Smith, B.J, Colman, P.M, Von Itzstein, M, Danylec, B, Varghese, J.N. | | Deposit date: | 2000-06-30 | | Release date: | 2001-04-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Analysis of inhibitor binding in influenza virus neuraminidase.
Protein Sci., 10, 2001
|
|
1F8E
 
 | | Native Influenza Neuraminidase in Complex with 4,9-diamino-2-deoxy-2,3-dehydro-N-acetyl-neuraminic Acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,9-AMINO-2,4-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, ... | | Authors: | Smith, B.J, Colman, P.M, Von Itzstein, M, Danylec, B, Varghese, J.N. | | Deposit date: | 2000-06-30 | | Release date: | 2001-04-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Analysis of inhibitor binding in influenza virus neuraminidase.
Protein Sci., 10, 2001
|
|
1USX
 
 | | Crystal structure of the Newcastle disease virus hemagglutinin-neuraminidase complexed with thiosialoside | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, HEMAGGLUTININ-NEURAMINIDASE GLYCOPROTEIN, N-acetyl-alpha-neuraminic acid-(2-6)-methyl 6-thio-beta-D-galactopyranoside | | Authors: | Zaitsev, V, Itzstein, M, Groves, D, Kiefel, M, Takimoto, T, Portner, A, Taylor, G. | | Deposit date: | 2003-12-01 | | Release date: | 2004-03-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Second Sialic Acid Binding Site in Newcastle Disease Virus Hemagglutinin-Neuraminidase: Implications for Fusion
J.Virol., 78, 2004
|
|
5OV7
 
 | | tubulin - rigosertib complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Menchon, G, Prota, A.E, Steinmetz, M, Jost, M. | | Deposit date: | 2017-08-28 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Combined CRISPRi/a-Based Chemical Genetic Screens Reveal that Rigosertib Is a Microtubule-Destabilizing Agent.
Mol. Cell, 68, 2017
|
|
6TXR
 
 | | Structural insights into cubane-modified aptamer recognition of a malaria biomarker | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Cheung, Y, Roethlisberger, P, Mechaly, A, Weber, P, Wong, A, Lo, Y, Haouz, A, Savage, P, Hollenstein, M, Tanner, J. | | Deposit date: | 2020-01-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of abiotic cubane chemistries in a nucleic acid aptamer allows selective recognition of a malaria biomarker.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7B6J
 
 | | Crystal structure of MurE from E.coli in complex with minifrag succinimide | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase, ... | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
7B6G
 
 | | Crystal structure of MurE from E.coli in complex with Z1675346324 | | Descriptor: | DIMETHYL SULFOXIDE, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase, trans-3-[(2,6-dimethylpyrimidin-4-yl)(methyl)amino]cyclobutan-1-ol | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
7B6Q
 
 | | Crystal structure of MurE from E.coli in complex with Z57299526 | | Descriptor: | ISOPROPYL ALCOHOL, N-[(furan-2-yl)methyl]-1H-benzimidazol-2-amine, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-08 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
7B68
 
 | | Crystal structure of MurE from E.coli in complex with Z57299526 | | Descriptor: | 4-[(4-methylphenyl)methyl]-1,4-thiazinane 1,1-dioxide, DIMETHYL SULFOXIDE, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
7B6M
 
 | | Crystal structure of MurE from E.coli in complex with Z1198948504 | | Descriptor: | 1,2-ETHANEDIOL, N-[2-(2,5-Dioxopyrrolidin-1-yl)ethyl]-3-methylbenzamide, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC), Structural Genomics Consortium for Research on Gene Expression (SGCGES) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
7B53
 
 | | Crystal structure of MurE from E.coli | | Descriptor: | 1,2-ETHANEDIOL, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
7B6N
 
 | | Crystal structure of MurE from E.coli in complex with Z757284952 | | Descriptor: | 1,2-ETHANEDIOL, N-(2-(2,4-dioxothiazolidin-3-yl)ethyl)-3-methylbenzamide, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
