3R5P
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824 | | Descriptor: | Deazaflavin-dependent nitroreductase, SULFATE ION | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-19 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5Y
 
 | | Structure of a Deazaflavin-dependent nitroreductase from Nocardia farcinica, with co-factor F420 | | Descriptor: | COENZYME F420, Putative uncharacterized protein | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5W
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824, with co-factor F420 | | Descriptor: | COENZYME F420, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
2HXZ
 
 | | Crystal Structure of Cathepsin S in complex with a Nonpeptidic Inhibitor (Hexagonal spacegroup) | | Descriptor: | Cathepsin S, N-[(1S)-1-{1-[(1R,3E)-1-ACETYLPENT-3-EN-1-YL]-1H-1,2,3-TRIAZOL-4-YL}-1,2-DIMETHYLPROPYL]BENZAMIDE, SULFATE ION | | Authors: | Patterson, A.W, Wood, W.J, Hornsby, M, Lesley, S, Spraggon, G, Ellman, J.A. | | Deposit date: | 2006-08-04 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of selective, nonpeptidic nitrile inhibitors of cathepsin s using the substrate activity screening method.
J.Med.Chem., 49, 2006
|
|
6UIP
 
 | | DYRK1A Kinase Domain in Complex with a 6-azaindole Derivative, GNF2133. | | Descriptor: | 4-ethyl-N-{4-[1-(oxan-4-yl)-1H-pyrrolo[2,3-c]pyridin-3-yl]pyridin-2-yl}piperazine-1-carboxamide, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2019-10-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Selective DYRK1A Inhibitor for the Treatment of Type 1 Diabetes: Discovery of 6-Azaindole Derivative GNF2133.
J.Med.Chem., 63, 2020
|
|
7RUN
 
 | | Crystal structure of phosphorylated RET tyrosine kinase domain complexed with a pyrrolo[2,3-d]pyrimidine inhibitor. | | Descriptor: | 1-(4-{(1s,3s)-3-[4-amino-5-(3-amino-4-chlorophenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl]cyclobutyl}piperazin-1-yl)ethan-1-one, CHLORIDE ION, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Lee, C.C, Spraggon, G. | | Deposit date: | 2021-08-17 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Antitarget Selectivity and Tolerability of Novel Pyrrolo[2,3- d ]pyrimidine RET Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
1S96
 
 | | The 2.0 A X-ray structure of Guanylate Kinase from E.coli | | Descriptor: | Guanylate kinase, PHOSPHATE ION, UNKNOWN ATOM OR ION | | Authors: | Kreusch, A, Spraggon, G, Klock, H.E, McMullan, D, Vincent, J, Rodrigues, K, Lesley, S.A. | | Deposit date: | 2004-02-03 | | Release date: | 2004-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of Guanylate Kinase from Escherichia coli at 2.0 A resolution
To be Published
|
|
4MKC
 
 | | Crystal Structure of Anaplastic Lymphoma Kinase Complexed with LDK378 | | Descriptor: | 5-chloro-N~2~-[5-methyl-4-(piperidin-4-yl)-2-(propan-2-yloxy)phenyl]-N~4~-[2-(propan-2-ylsulfonyl)phenyl]pyrimidine-2,4-diamine, ALK tyrosine kinase receptor, GLYCEROL | | Authors: | Lee, C.C, Spraggon, G. | | Deposit date: | 2013-09-04 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The ALK Inhibitor Ceritinib Overcomes Crizotinib Resistance in Non-Small Cell Lung Cancer.
Cancer Discov, 4, 2014
|
|
1J7J
 
 | |
1FZE
 
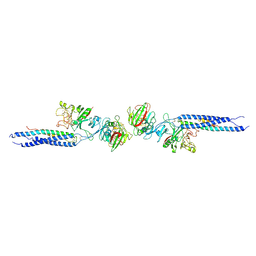 | | CRYSTAL STRUCTURE OF FRAGMENT DOUBLE-D FROM HUMAN FIBRIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FIBRINOGEN | | Authors: | Everse, S.J, Spraggon, G, Veerapandian, L, Doolittle, R.F. | | Deposit date: | 1998-12-23 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Conformational changes in fragments D and double-D from human fibrin(ogen) upon binding the peptide ligand Gly-His-Arg-Pro-amide.
Biochemistry, 38, 1999
|
|
1FZF
 
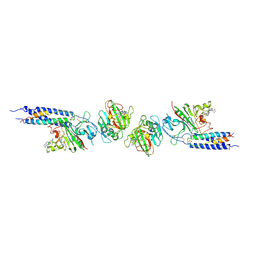 | | CRYSTAL STRUCTURE OF FRAGMENT DOUBLE-D FROM HUMAN FIBRIN WITH THE PEPTIDE LIGAND GLY-HIS-ARG-PRO-AMIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FIBRINOGEN | | Authors: | Everse, S.J, Spraggon, G, Veerapandian, L, Doolittle, R.F. | | Deposit date: | 1998-12-28 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational changes in fragments D and double-D from human fibrin(ogen) upon binding the peptide ligand Gly-His-Arg-Pro-amide.
Biochemistry, 38, 1999
|
|
1FZG
 
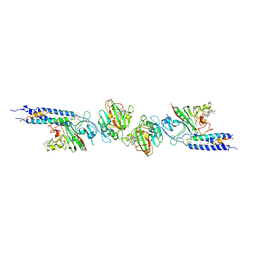 | | CRYSTAL STRUCTURE OF FRAGMENT D FROM HUMAN FIBRINOGEN WITH THE PEPTIDE LIGAND GLY-HIS-ARG-PRO-AMIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FIBRINOGEN | | Authors: | Everse, S.J, Spraggon, G, Veerapandian, L, Doolittle, R.F. | | Deposit date: | 1999-01-01 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational changes in fragments D and double-D from human fibrin(ogen) upon binding the peptide ligand Gly-His-Arg-Pro-amide.
Biochemistry, 38, 1999
|
|
1LWU
 
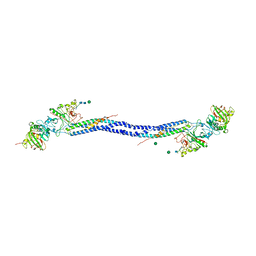 | | Crystal structure of fragment D from lamprey fibrinogen complexed with the peptide Gly-His-Arg-Pro-amide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, Z, Spraggon, G, Pandi, L, Everse, S.J, Riley, M, Doolittle, R.F. | | Deposit date: | 2002-06-03 | | Release date: | 2002-08-23 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of fragment D from lamprey fibrinogen complexed with the peptide Gly-His-Arg-Pro-amide.
Biochemistry, 41, 2002
|
|
1LM4
 
 | | Structure of Peptide Deformylase from Staphylococcus aureus at 1.45 A | | Descriptor: | FE (III) ION, GLYCEROL, peptide deformylase PDF1 | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A. | | Deposit date: | 2002-04-30 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
1LME
 
 | | Crystal Structure of Peptide Deformylase from Thermotoga maritima | | Descriptor: | peptide deformylase | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2002-05-01 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
1LM6
 
 | | Crystal Structure of Peptide Deformylase from Streptococcus pneumoniae | | Descriptor: | FE (III) ION, GLYCEROL, peptide deformylase DEFB | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A. | | Deposit date: | 2002-04-30 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
1N5N
 
 | | Crystal Structure of Peptide Deformylase from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, Peptide deformylase, ZINC ION | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A. | | Deposit date: | 2002-11-06 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
1KGZ
 
 | | Crystal Structure Analysis of the Anthranilate Phosphoribosyltransferase from Erwinia carotovora (current name, Pectobacterium carotovorum) | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Anthranilate phosphoribosyltransferase, MANGANESE (II) ION, ... | | Authors: | Kim, C, Xuong, N.-H, Edwards, S, Madhusudan, Yee, M.-C, Spraggon, G, Mills, S.E. | | Deposit date: | 2001-11-28 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of Anthranilate
Phosphoribosyltransferase from the
Enterobacterium Pectobacterium carotovorum
FEBS Lett., 523, 2002
|
|
1NH1
 
 | | Crystal Structure of the Type III Effector AvrB from Pseudomonas syringae. | | Descriptor: | Avirulence B protein | | Authors: | Lee, C.C, Wood, M.D, Ng, K, Luginbuhl, P, Spraggon, G, Katagiri, F. | | Deposit date: | 2002-12-18 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Type III Effector AvrB from Pseudomonas syringae.
Structure, 12, 2004
|
|
1KEX
 
 | | Crystal Structure of the b1 Domain of Human Neuropilin-1 | | Descriptor: | Neuropilin-1 | | Authors: | Lee, C.C, Kreusch, A, McMullan, D, Ng, K, Spraggon, G. | | Deposit date: | 2001-11-18 | | Release date: | 2003-01-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Human Neuropilin-1 b1 Domain
Structure, 11, 2003
|
|
1KHD
 
 | | Crystal Structure Analysis of the anthranilate phosphoribosyltransferase from Erwinia carotovora at 1.9 resolution (current name, Pectobacterium carotovorum) | | Descriptor: | Anthranilate phosphoribosyltransferase | | Authors: | Kim, C, Xuong, N.-H, Edwards, S, Madhusudan, Yee, M.-C, Spraggon, G, Mills, S.E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-10-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Crystal Structure of Anthranilate
Phosphoribosyltransferase from the
Enterobacterium Pectobacterium carotovorum
FEBS Lett., 523, 2002
|
|
4Z55
 
 | |
6VHG
 
 | | Crystal structure of phosphorylated RET tyrosine kinase domain complexed with a pyrazolo[1,5-a]pyrimidine inhibitor | | Descriptor: | 3-(3,4-dimethoxyphenyl)-N~5~-(1-methylpiperidin-4-yl)-6-phenylpyrazolo[1,5-a]pyrimidine-5,7-diamine, ACETATE ION, CHLORIDE ION, ... | | Authors: | Lee, C.C, Spraggon, G. | | Deposit date: | 2020-01-09 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Efficacy and Tolerability of Pyrazolo[1,5-a]pyrimidine RET Kinase Inhibitors for the Treatment of Lung Adenocarcinoma.
Acs Med.Chem.Lett., 11, 2020
|
|
3O0P
 
 | | Pilus-related Sortase C of Group B Streptococcus | | Descriptor: | Sortase family protein | | Authors: | Malito, E, Spraggon, G. | | Deposit date: | 2010-07-19 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure analysis and site-directed mutagenesis of defined key residues and motives for pilus-related sortase C1 in group B Streptococcus.
Faseb J., 25, 2011
|
|
2Y7S
 
 | |
