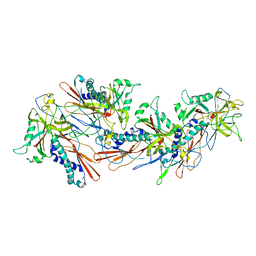
1G4B
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | Descriptor: | ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
1G4A
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
8T1U
 
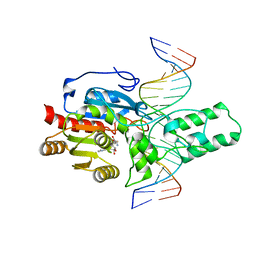 | | Crystal structure of the DRM2-CTA DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*TP*AP*TP*TP*AP*AP*TP*(C49)P*TP*AP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(P*TP*AP*AP*AP*TP*TP*TP*AP*GP*AP*TP*TP*AP*AP*TP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Chen, J, Lu, J, Song, J. | | Deposit date: | 2023-06-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | DNA conformational dynamics in the context-dependent non-CG CHH methylation by plant methyltransferase DRM2.
J.Biol.Chem., 299, 2023
|
|
8T12
 
 | |
8T13
 
 | |
6P3W
 
 | |
8TCI
 
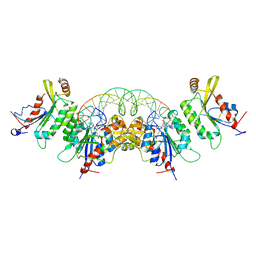 | | Crystal structure of DNMT3C-DNMT3L in complex with CGG DNA | | Descriptor: | (5'-D(P*CP*AP*TP*G)-R(P*(PYO))-D(P*GP*GP*TP*CP*TP*AP*AP*TP*TP*AP*GP*AP*CP*CP*GP*CP*AP*TP*G)-3'), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3C, ... | | Authors: | Khudaverdyan, N, Song, J. | | Deposit date: | 2023-07-01 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The structure of DNA methyltransferase DNMT3C reveals an activity-tuning mechanism for DNA methylation.
J.Biol.Chem., 300, 2024
|
|
5TMH
 
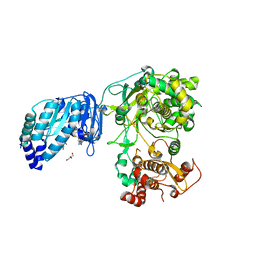 | | Structure of Zika virus NS5 | | Descriptor: | GLYCEROL, Polyprotein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Wang, B, Tan, X, Song, J. | | Deposit date: | 2016-10-12 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.278 Å) | | Cite: | The structure of Zika virus NS5 reveals a conserved domain conformation.
Nat Commun, 8, 2017
|
|
3GXU
 
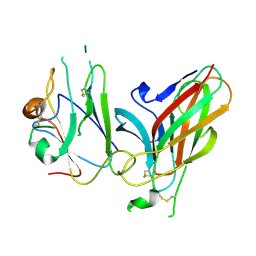 | | Crystal structure of Eph receptor and ephrin complex | | Descriptor: | Ephrin type-A receptor 4, Ephrin-B2 | | Authors: | Qin, H.N, Song, J.X. | | Deposit date: | 2009-04-03 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural characterization of the EphA4-ephrin-B2 complex reveals new features enabling Eph-ephrin binding promiscuity
J.Biol.Chem., 2009
|
|
7LMM
 
 | |
7LMK
 
 | |
7L4C
 
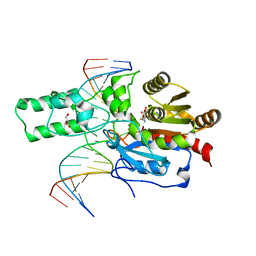 | | Crystal structure of the DRM2-CTT DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*TP*TP*AP*AP*TP*(C49)P*TP*TP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*AP*AP*GP*AP*TP*TP*AP*AP*TP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-18 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4K
 
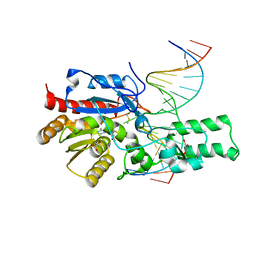 | | Crystal structure of the DRM2-CCG DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*CP*TP*AP*AP*TP*(C49)P*CP*GP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*CP*GP*GP*AP*TP*TP*AP*GP*GP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4H
 
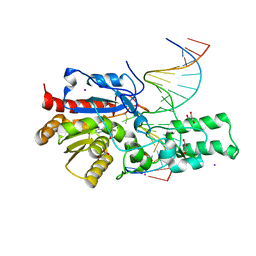 | | Crystal structure of the DRM2-CTG DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*CP*TP*AP*AP*TP*(C49)P*TP*GP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*CP*AP*GP*AP*TP*TP*AP*GP*GP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4N
 
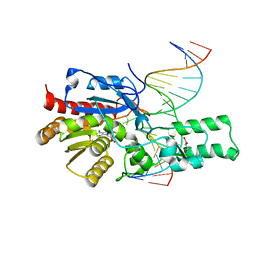 | | Crystal structure of the DRM2 (C397R)-CCG DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*CP*TP*AP*AP*TP*(C49)P*CP*GP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*CP*GP*GP*AP*TP*TP*AP*GP*GP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4M
 
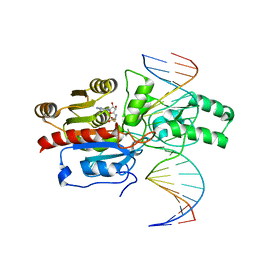 | | Crystal structure of the DRM2-CCT DNA complex | | Descriptor: | DNA (5'-D(*TP*AP*AP*AP*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*AP*AP*T)-3'), DNA (5'-D(P*AP*TP*TP*CP*CP*TP*CP*CP*TP*(C49)P*CP*TP*CP*CP*TP*TP*TP*A)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4F
 
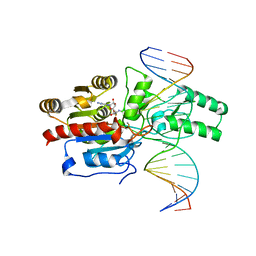 | | Crystal structure of the DRM2-CAT DNA complex | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*AP*TP*TP*CP*CP*TP*CP*CP*TP*(C49)P*AP*TP*CP*CP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*GP*GP*AP*TP*GP*AP*GP*GP*AP*GP*GP*AP*AP*T)-3'), ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
8FZB
 
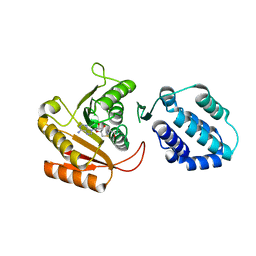 | | Crystal structure of human FAM86A | | Descriptor: | Protein-lysine N-methyltransferase EEF2KMT, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Shao, Z, Lu, J, Song, J. | | Deposit date: | 2023-01-28 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | The FAM86 domain of FAM86A confers substrate specificity to promote EEF2-Lys525 methylation.
J.Biol.Chem., 299, 2023
|
|
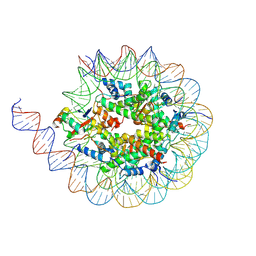
8GN5
 
 | | Designed pH-responsive P22 VLP | | Descriptor: | Major capsid protein | | Authors: | Kim, K.J, Kim, G, Bae, J.H, Song, J.J, Kim, H.S. | | Deposit date: | 2022-08-23 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | A pH-Responsive Virus-Like Particle as a Protein Cage for a Targeted Delivery.
Adv Healthc Mater, 13, 2024
|
|
8U5H
 
 | |
7UBU
 
 | |
8FZ9
 
 | |
1PW4
 
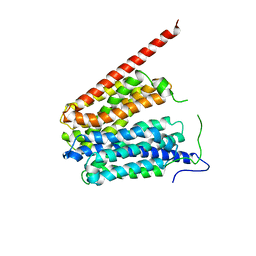 | | Crystal Structure of the Glycerol-3-Phosphate Transporter from E.Coli | | Descriptor: | Glycerol-3-phosphate transporter | | Authors: | Huang, Y, Lemieux, M.J, Song, J, Auer, M, Wang, D.N. | | Deposit date: | 2003-06-30 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and Mechanism of the Glycerol-3-Phosphate Transporter from Escherichia Coli
Science, 301, 2003
|
|
6UCA
 
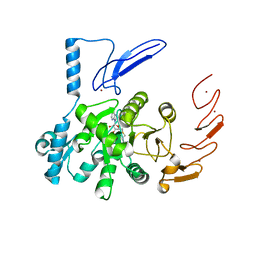 | | Crystal structure of human ZCCHC4 in complex with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, rRNA N6-adenosine-methyltransferase ZCCHC4 | | Authors: | Lu, J.W, Ren, W.D, Huang, M.J, Gao, L, Li, D.X, Wang, G.G, Song, J. | | Deposit date: | 2019-09-15 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structure and regulation of ZCCHC4 in m6A-methylation of 28S rRNA.
Nat Commun, 10, 2019
|
|
6AGO
 
 | | Crystal structure of MRG15-ASH1L Histone methyltransferase complex | | Descriptor: | Histone-lysine N-methyltransferase ASH1L, Mortality factor 4 like 1, S-ADENOSYLMETHIONINE, ... | | Authors: | Lee, Y, Song, J. | | Deposit date: | 2018-08-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structural Basis of MRG15-Mediated Activation of the ASH1L Histone Methyltransferase by Releasing an Autoinhibitory Loop.
Structure, 27, 2019
|
|
