3NNH
 
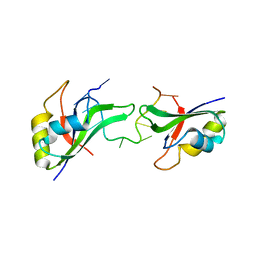 | | Crystal Structure of the CUGBP1 RRM1 with GUUGUUUUGUUU RNA | | Descriptor: | CUGBP Elav-like family member 1, RNA (5'-R(*GP*UP*UP*GP*UP*UP*UP*UP*GP*UP*UP*U)-3') | | Authors: | Teplova, M, Song, J, Gaw, H, Teplov, A, Patel, D.J. | | Deposit date: | 2010-06-23 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7501 Å) | | Cite: | Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1.
Structure, 18, 2010
|
|
4ET7
 
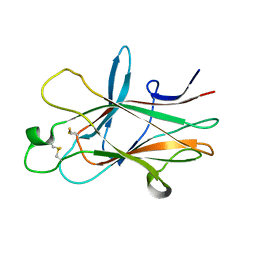 | |
3OPE
 
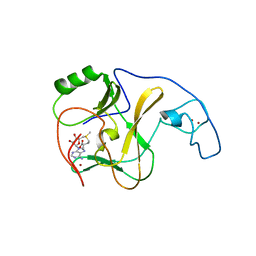 | | Structural Basis of Auto-inhibitory mechanism of Histone methyltransferase | | Descriptor: | Probable histone-lysine N-methyltransferase ASH1L, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | An, S, Song, J. | | Deposit date: | 2010-08-31 | | Release date: | 2011-01-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the human histone methyltransferase ASH1L catalytic domain and its implications for the regulatory mechanism
J.Biol.Chem., 286, 2011
|
|
2GJI
 
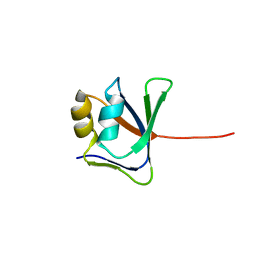 | | NMR solution structure of VP9 from White Spot Syndrome Virus | | Descriptor: | wsv230 | | Authors: | Liu, Y, Wu, J.L, Song, J.X, Sivaraman, J, Hew, C.L. | | Deposit date: | 2006-03-30 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Identification of a Novel Nonstructural Protein, VP9, from White Spot Syndrome Virus: Its Structure Reveals a Ferredoxin Fold with Specific Metal Binding Sites
J.Virol., 80, 2006
|
|
3NNA
 
 | | Crystal Structure of CUGBP1 RRM1/2-RNA Complex | | Descriptor: | CUGBP Elav-like family member 1, RNA (5'-R(*GP*UP*UP*GP*UP*UP*UP*UP*GP*UP*UP*U)-3') | | Authors: | Teplova, M, Song, J, Gaw, H, Teplov, A, Patel, D.J. | | Deposit date: | 2010-06-23 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1.
Structure, 18, 2010
|
|
3W5K
 
 | | Crystal structure of Snail1 and importin beta complex | | Descriptor: | Importin subunit beta-1, ZINC ION, Zinc finger protein SNAI1 | | Authors: | Choi, S, Yamashita, E, Yasuhara, N, Song, J, Son, S.Y, Won, Y.H, Shin, Y.S, Sekimoto, T, Park, I.Y, Yoneda, Y, Lee, S.J. | | Deposit date: | 2013-01-30 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the selective nuclear import of the C2H2 zinc-finger protein Snail by importin beta.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5YX2
 
 | |
2OP7
 
 | | WW4 | | Descriptor: | NEDD4-like E3 ubiquitin-protein ligase WWP1 | | Authors: | Qin, H.N, Li, M.F, Pu, H, Sankaran, S, Ahmed, S, Song, J.X. | | Deposit date: | 2007-01-27 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the forth WW domain of WWP1
To be Published
|
|
4ZIR
 
 | | Crystal structure of EcfAA' heterodimer bound to AMPPNP | | Descriptor: | CHLORIDE ION, Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, ... | | Authors: | Karpowich, N.K, Cocco, N, Song, J.M, Wang, D.N. | | Deposit date: | 2015-04-28 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | ATP binding drives substrate capture in an ECF transporter by a release-and-catch mechanism.
Nat.Struct.Mol.Biol., 22, 2015
|
|
8TE4
 
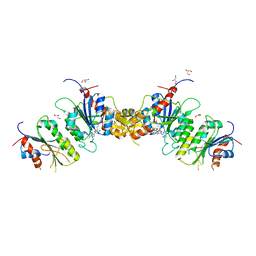 | |
8TE3
 
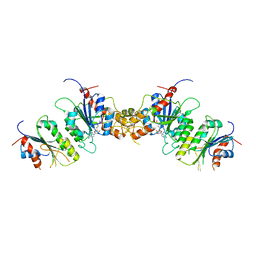 | |
8TDR
 
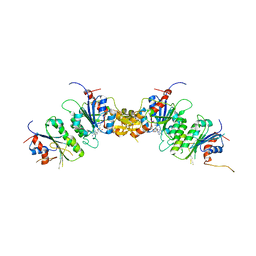 | |
8TE1
 
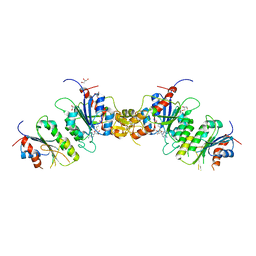 | |
4LUB
 
 | |
2G31
 
 | | Human Nogo-A functional domain: nogo60 | | Descriptor: | Reticulon-4 | | Authors: | Li, M.F, Liu, J.X, Song, J.X. | | Deposit date: | 2006-02-17 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nogo goes in the pure water: solution structure of Nogo-60 and design of the structured and buffer-soluble Nogo-54 for enhancing CNS regeneration
Protein Sci., 15, 2006
|
|
2FB7
 
 | |
2I85
 
 | |
8T1U
 
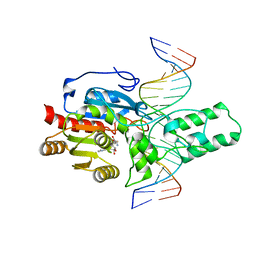 | | Crystal structure of the DRM2-CTA DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*TP*AP*TP*TP*AP*AP*TP*(C49)P*TP*AP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(P*TP*AP*AP*AP*TP*TP*TP*AP*GP*AP*TP*TP*AP*AP*TP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Chen, J, Lu, J, Song, J. | | Deposit date: | 2023-06-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | DNA conformational dynamics in the context-dependent non-CG CHH methylation by plant methyltransferase DRM2.
J.Biol.Chem., 299, 2023
|
|
8T12
 
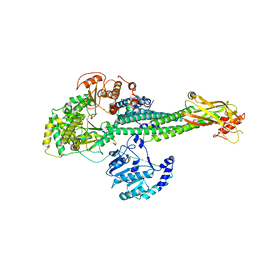 | |
8T13
 
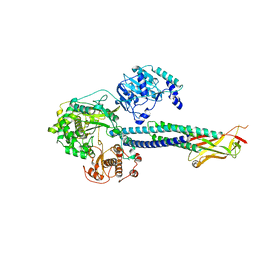 | |
5KBW
 
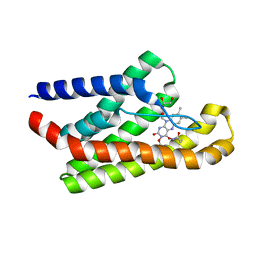 | |
5KC4
 
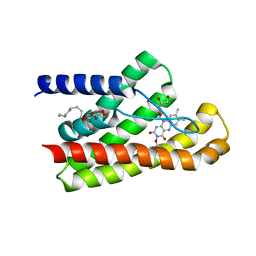 | | Structure of TmRibU, orthorhombic crystal form | | Descriptor: | RIBOFLAVIN, Riboflavin transporter RibU, nonyl beta-D-glucopyranoside | | Authors: | Karpowich, N.K, Wang, D.N, Song, J.M. | | Deposit date: | 2016-06-04 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | An Aromatic Cap Seals the Substrate Binding Site in an ECF-Type S Subunit for Riboflavin.
J.Mol.Biol., 428, 2016
|
|
5KC0
 
 | | Crystal structure of TmRibU, hexagonal crystal form | | Descriptor: | RIBOFLAVIN, Riboflavin transporter RibU, nonyl beta-D-glucopyranoside | | Authors: | Karpowich, N.K, Wang, D.N, Song, J.M. | | Deposit date: | 2016-06-03 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2001 Å) | | Cite: | An Aromatic Cap Seals the Substrate Binding Site in an ECF-Type S Subunit for Riboflavin.
J.Mol.Biol., 428, 2016
|
|
6B3X
 
 | | Crystal structure of CstF-50 in complex with CstF-77 | | Descriptor: | Cleavage stimulation factor subunit 1, Cleavage stimulation factor subunit 3 | | Authors: | Yang, W, Hsu, P, Yang, F, Song, J.E, Varani, G. | | Deposit date: | 2017-09-25 | | Release date: | 2017-11-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reconstitution of the CstF complex unveils a regulatory role for CstF-50 in recognition of 3'-end processing signals.
Nucleic Acids Res., 46, 2018
|
|
4Z97
 
 | |
