8PNR
 
 | | Structure of human KCTD15 BTB domain mutant G88D crystal form 1 | | Descriptor: | BTB/POZ domain-containing protein KCTD15 | | Authors: | Cruz Walma, D.A, Cros, J, Bradshaw, W, Richardson, W, Chen, Z, Chalk, R, Wilkie, A, Bullock, A.N. | | Deposit date: | 2023-06-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | BTB domain mutations perturbing KCTD15 oligomerisation cause a distinctive frontonasal dysplasia syndrome.
J Med Genet, 61, 2024
|
|
8PNM
 
 | | Structure of human KCTD15 BTB domain mutant G88D crystal form 2 | | Descriptor: | BTB/POZ domain-containing protein KCTD15 | | Authors: | Cruz Walma, D.A, Cros, J, Bradshaw, W, Richardson, W, Chen, Z, Chalk, R, Wilkie, A, Bullock, A.N. | | Deposit date: | 2023-06-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | BTB domain mutations perturbing KCTD15 oligomerisation cause a distinctive frontonasal dysplasia syndrome.
J Med Genet, 61, 2024
|
|
8HPI
 
 | |
8HPG
 
 | |
8I7H
 
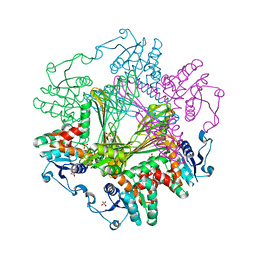 | | Meso-Diaminopimelate dehydrogenase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, Meso-diaminopimelate D-dehydrogenase, SULFATE ION | | Authors: | Wu, T.F, Song, W. | | Deposit date: | 2023-01-31 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Analysis of the catalytic mechanism of meso-DAPDH and extension of D-aromatic amino acid substrate scope
To Be Published
|
|
3IB3
 
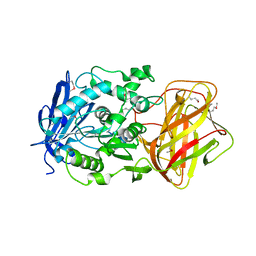 | | Crystal Structure of SACOL2612 - CocE/NonD family hydrolase from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, CocE/NonD family hydrolase, NICKEL (II) ION, ... | | Authors: | Domagalski, M.J, Chruszcz, M, Osinski, T, Skarina, T, Onopriyenko, O, Cymborowski, M, Shumilin, I.A, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-15 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of SACOL2612 - CocE/NonD family hydrolase from Staphylococcus aureus
To be Published
|
|
8HPC
 
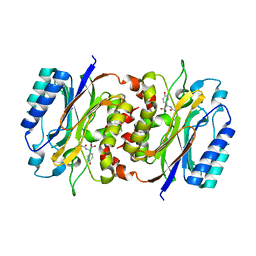 | |
8HP4
 
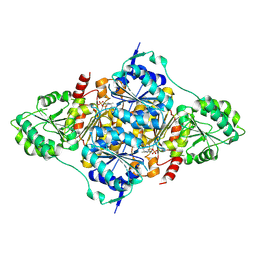 | | CtPDC complex | | Descriptor: | MAGNESIUM ION, Pyruvate decarboxylase, THIAMINE DIPHOSPHATE | | Authors: | Xu, H.H, Song, W. | | Deposit date: | 2022-12-11 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Protein engineering of pyruvate decarboxylase to remove the rate-limiting bottleneck in the cascade pathway of tyrosol synthesis
To Be Published
|
|
8HP2
 
 | | CtPDC | | Descriptor: | Pyruvate decarboxylase | | Authors: | Xu, H.H, Song, W. | | Deposit date: | 2022-12-11 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Protein engineering of pyruvate decarboxylase to remove the rate-limiting bottleneck in the cascade pathway of tyrosol synthesis
To Be Published
|
|
3SKS
 
 | | Crystal structure of a putative oligoendopeptidase F from Bacillus anthracis str. Ames | | Descriptor: | PHOSPHATE ION, Putative Oligoendopeptidase F, ZINC ION | | Authors: | Wajerowicz, W, Onopriyenko, O, Porebski, P, Domagalski, M, Chruszcz, M, Savchenko, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-23 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a putative oligoendopeptidase F from Bacillus anthracis str. Ames
TO BE PUBLISHED
|
|
5XLR
 
 | | Structure of SARS-CoV spike glycoprotein | | Descriptor: | Spike glycoprotein | | Authors: | Gui, M, Song, W, Xiang, Y, Wang, X. | | Deposit date: | 2017-05-11 | | Release date: | 2017-06-07 | | Last modified: | 2019-10-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding.
Cell Res., 27, 2017
|
|
8OIO
 
 | | Crystal structure of the kelch domain of human KLHL12 in complex with PLEKHA4 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Kelch-like protein 12, ... | | Authors: | Dalietou, E.V, Chen, Z, Ramdass, A.E, Manning, C, Richardson, W, Aitmakhanova, K, Platt, M, Pike, A.C.W, Fedorov, O, Brennan, P, Bullock, A.N. | | Deposit date: | 2023-03-23 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Crystal structure of the kelch domain of human KLHL12 in complex with PLEKHA4 peptide
To Be Published
|
|
6ACG
 
 | |
6ACJ
 
 | |
8R5J
 
 | | Crystal structure of MERS-CoV main protease | | Descriptor: | Non-structural protein 11 | | Authors: | Balcomb, B.H, Fairhead, M, Koekemoer, L, Lithgo, R.M, Aschenbrenner, J.C, Chandran, A.V, Godoy, A.S, Lukacik, P, Marples, P.G, Mazzorana, M, Ni, X, Strain-Damerell, C, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-11-16 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal structure of MERS-CoV main protease
To Be Published
|
|
5BJO
 
 | | Crystal structure of the Corn RNA aptamer in complex with DFHO, site-specific 5-iodo-U | | Descriptor: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Warner, K.D, Song, W, Filonov, G.S, Jaffrey, S.R, Ferre-D'Amare, A.R. | | Deposit date: | 2016-08-21 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A homodimer interface without base pairs in an RNA mimic of red fluorescent protein.
Nat. Chem. Biol., 13, 2017
|
|
5BJP
 
 | | Crystal structure of the Corn RNA aptamer in complex with DFHO, iridium hexammine soak | | Descriptor: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, DIMETHYL SULFOXIDE, IRIDIUM ION, ... | | Authors: | Warner, K.D, Song, W, Filonov, G.S, Jaffrey, S.R, Ferre-D'Amare, A.R. | | Deposit date: | 2016-08-21 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | A homodimer interface without base pairs in an RNA mimic of red fluorescent protein.
Nat. Chem. Biol., 13, 2017
|
|
7O2T
 
 | | PqsR (MvfR) in complex with antagonist 61 | | Descriptor: | 2-[4-[(2S)-3-(6-chloro-4-oxoquinazolin-3-yl)-2-hydroxypropoxy]phenyl]acetonitrile, LysR family transcriptional regulator | | Authors: | Emsley, J, Richardson, W. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | PqsR (MvfR) in complex with antagonist 61
To Be Published
|
|
7O2U
 
 | | PqsR (MvfR) in complex with antagonist 40 | | Descriptor: | 2-[4-[(2S)-3-(6-chloranyl-4-oxidanylidene-quinazolin-3-yl)-2-oxidanyl-propoxy]phenoxy]ethanenitrile, LysR family transcriptional regulator | | Authors: | Emsley, J, Richardson, W. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | PqsR (MvfR) in complex with antagonist 40
To Be Published
|
|
3P7M
 
 | | Structure of putative lactate dehydrogenase from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | Malate dehydrogenase, PHOSPHATE ION | | Authors: | Osinski, T, Cymborowski, M, Zimmerman, M.D, Gordon, E, Grimshaw, S, Skarina, T, Chruszcz, M, Savchenko, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-12 | | Release date: | 2010-10-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of putative lactate dehydrogenase from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
3SLF
 
 | | Crystal structure of BA2930 in complex with AcCoA and uracil | | Descriptor: | ACETYL COENZYME *A, Aminoglycoside N3-acetyltransferase, CHLORIDE ION, ... | | Authors: | Klimecka, M.M, Chruszcz, M, Porebski, P.J, Cymborowski, M, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-24 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of BA2930 in complex with AcCoA and uracil
TO BE PUBLISHED
|
|
3SLB
 
 | | Crystal structure of BA2930 in complex with AcCoA and cytosine | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 6-AMINOPYRIMIDIN-2(1H)-ONE, ... | | Authors: | Klimecka, M.M, Chruszcz, M, Porebski, P.J, Cymborowski, M, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-24 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of BA2930 in complex with AcCoA and cytosine
TO BE PUBLISHED
|
|
3EFE
 
 | | The crystal structure of the thiJ/pfpI family protein from Bacillus anthracis | | Descriptor: | SULFATE ION, ThiJ/pfpI family protein | | Authors: | Zhang, R, Xu, X, Cui, H, Savchenko, A, Edwards, A, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-08 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the thiJ/pfpI family protein from Bacillus anthracis
To be Published
|
|
3E7N
 
 | | Crystal structure of d-ribose high-affinity transport system from salmonella typhimurium lt2 | | Descriptor: | 1,2-ETHANEDIOL, D-ribose high-affinity transport system | | Authors: | Nocek, B, Maltseva, N, Gu, M, Joachimiak, A, Anderson, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-08-18 | | Release date: | 2008-08-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of d-ribose high-affinity transport system from salmonella typhimurium lt2
To be Published
|
|
3CWC
 
 | | Crystal structure of putative glycerate kinase 2 from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative glycerate kinase 2 | | Authors: | Osipiuk, J, Zhou, M, Holzle, D, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-04-21 | | Release date: | 2008-07-08 | | Last modified: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | X-ray crystal structure of putative glycerate kinase 2 from Salmonella typhimurium LT2.
To be Published
|
|
