3LIB
 
 | |
4R57
 
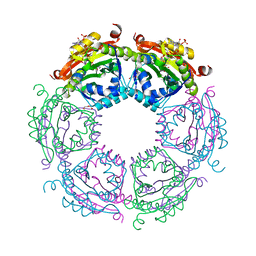 | | Crystal structure of spermidine N-acetyltransferase from Vibrio cholerae in complex with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, DI(HYDROXYETHYL)ETHER, Spermidine n1-acetyltransferase, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Kuhn, M.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-20 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | A Novel Polyamine Allosteric Site of SpeG from Vibrio cholerae Is Revealed by Its Dodecameric Structure.
J.Mol.Biol., 427, 2015
|
|
6IWY
 
 | |
8T4N
 
 | |
3MJZ
 
 | | The crystal structure of native FG41 MSAD | | Descriptor: | FG41 Malonate Semialdehyde Decarboxylase | | Authors: | Guo, Y, Serrano, H, Poelarends, G.J, Johnson, W.H.Jr, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-04-13 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic, Mutational, and Structural Analysis of Malonate Semialdehyde Decarboxylase from Coryneform Bacterium Strain FG41: Mechanistic Implications for the Decarboxylase and Hydratase Activities.
Biochemistry, 52, 2013
|
|
8HP3
 
 | | Crystal structure of meso-diaminopimelate dehydrogenase from Prevotella timonensis | | Descriptor: | 1,2-ETHANEDIOL, Meso-diaminopimelate D-dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tan, Y, Song, W. | | Deposit date: | 2022-12-11 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Rational Design of Meso -Diaminopimelate Dehydrogenase with Enhanced Reductive Amination Activity for Efficient Production of d- p -Hydroxyphenylglycine.
Appl.Environ.Microbiol., 89, 2023
|
|
8HPE
 
 | | Crystal structure of Leucine dehydrogenase | | Descriptor: | GLYCEROL, Leucine dehydrogenase, SULFATE ION | | Authors: | Li, X, Song, W. | | Deposit date: | 2022-12-12 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | A Tri-Enzyme Cascade for Efficient Production of L-2-Aminobutyrate from L-Threonine.
Chembiochem, 24, 2023
|
|
8I1O
 
 | |
8I1N
 
 | |
6JV6
 
 | | Crystal structure of the sirohydrochlorin chelatase SirB from Bacillus subtilis subspecies spizizenii in complex with cobalt | | Descriptor: | COBALT (II) ION, Sirohydrochlorin ferrochelatase | | Authors: | Nam, M.S, Song, W.S, Park, S.C, Yoon, S.I. | | Deposit date: | 2019-04-16 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Cobalt complex structure of the sirohydrochlorin chelatase SirB from Bacillus subtilis subsp. spizizenii.
KOREAN J MICROBIOL., 55, 2019
|
|
6JYI
 
 | | Crystal structure of the PadR-like transcriptional regulator BC1756 from Bacillus cereus | | Descriptor: | Transcriptional repressor PadR | | Authors: | Kim, T.H, Park, S.C, Lee, K.C, Song, W.S, Yoon, S.I. | | Deposit date: | 2019-04-26 | | Release date: | 2019-06-26 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural and DNA-binding studies of the PadR-like transcriptional regulator BC1756 from Bacillus cereus.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
3OYT
 
 | | 1.84 Angstrom resolution crystal structure of 3-oxoacyl-(acyl carrier protein) synthase I (fabB) from Yersinia pestis CO92 | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier-protein] synthase I, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Onopriyenko, O, Peterson, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-23 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | 1.84 Angstrom resolution crystal structure of 3-oxoacyl-(acyl carrier protein) synthase I (fabB) from Yersinia pestis CO92
To be Published
|
|
2B0K
 
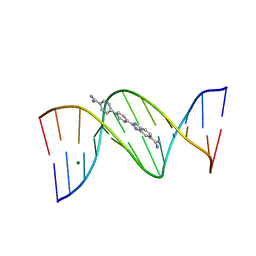 | | Crystal structure of the DB921-D(CGCGAATTCGCG)2 complex. | | Descriptor: | 2-(4'-AMIDINOBIPHENYL-4-YL)-1H-BENZIMIDAZOLE-5-AMIDINE, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Miao, Y, Lee, M.P, Parkinson, G.N, Batista-Parra, A, Ismail, M.A, Neidle, S, Boykin, D.W, Wilson, W.D. | | Deposit date: | 2005-09-14 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Out-of-Shape DNA Minor Groove Binders: Induced Fit Interactions of Heterocyclic Dications with the DNA Minor Groove.
Biochemistry, 44, 2005
|
|
4QJ1
 
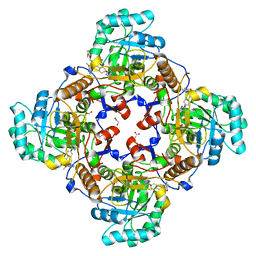 | | Co-crystal structure of the catalytic domain of the inosine monophosphate dehydrogenase from Cryptosporidium parvum with inhibitor N109 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, FORMIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-03 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.415 Å) | | Cite: | Co-crystal structure of the catalytic domain of the inosine monophosphate dehydrogenase from Cryptosporidium parvum with inhibitor N109
To be Published
|
|
4QVS
 
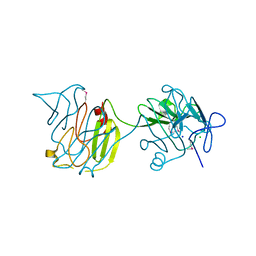 | | 2.1 Angstrom resolution crystal structure of S-layer domain-containing protein (residues 221-444) from Clostridium thermocellum ATCC 27405 | | Descriptor: | CHLORIDE ION, S-layer domain-containing protein, SODIUM ION | | Authors: | Halavaty, A.S, Wawrzak, Z, Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-15 | | Release date: | 2014-07-30 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom resolution crystal structure of S-layer domain-containing protein (residues 221-444) from Clostridium thermocellum ATCC 27405
To be Published
|
|
4R7K
 
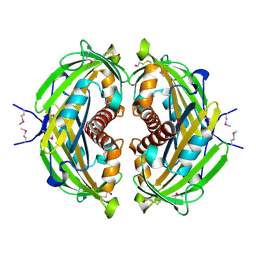 | | 1.88 Angstrom Resolution Crystal Structure of Hypothetical Protein jhp0584 from Helicobacter pylori. | | Descriptor: | Hypothetical protein jhp0584 | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Flores, K, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88 Angstrom Resolution Crystal Structure of Hypothetical Protein jhp0584 from Helicobacter pylori.
To be Published
|
|
4QRI
 
 | | 2.35 Angstrom resolution crystal structure of hypoxanthine-guanine-xanthine phosphoribosyltransferase from Leptospira interrogans serovar Copenhageni str. Fiocruz L1-130 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Hypoxanthine-guanine-xanthine phosphoribosyltransferase, SULFATE ION | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Flores, K.J, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-01 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom resolution crystal structure of hypoxanthine-guanine-xanthine phosphoribosyltransferase from Leptospira interrogans serovar Copenhageni str. Fiocruz L1-130
To be Published
|
|
4QUS
 
 | | 1.28 Angstrom resolution crystal structure of predicted acyltransferase with acyl-CoA N-acyltransferase domain (ypeA) from Escherichia coli str. K-12 substr. MG1655 | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase YpeA, PHOSPHATE ION | | Authors: | Halavaty, A.S, Filippova, E.V, Minasov, G, Flores, K.J, Dubrovska, I, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-11 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | 1.28 Angstrom resolution crystal structure of predicted acyltransferase with acyl-CoA N-acyltransferase domain (ypeA) from Escherichia coli str. K-12 substr. MG1655
To be Published
|
|
3TZC
 
 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG)(Y155F) from Vibrio cholerae | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Hou, J, Chruszcz, M, Zheng, H, Grabowski, M, Domagalski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-27 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
4R86
 
 | | Crystal Structure of Aminoglycoside/Multidrug Efflux System AcrD from Salmonella typhimurium | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Maltseva, N, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-29 | | Release date: | 2014-10-08 | | Last modified: | 2016-10-12 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Crystal Structure of Aminoglycoside/Multidrug Efflux System AcrD from
Salmonella typhimurium
To be Published
|
|
4RCO
 
 | | 1.9 Angstrom Crystal Structure of Superantigen-like Protein, Exotoxin from Staphylococcus aureus, in Complex with Sialyl-LewisX. | | Descriptor: | CHLORIDE ION, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose, Putative uncharacterized protein | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Filippova, E, Halavaty, A, Dubrovska, I, Flores, K, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-16 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Crystal Structure of Superantigen-like Protein, Exotoxin from Staphylococcus aureus, in Complex with Sialyl-LewisX.
TO BE PUBLISHED
|
|
4R9O
 
 | | Crystal Structure of Putative Aldo/Keto Reductase from Salmonella enterica | | Descriptor: | Putative aldo/keto reductase | | Authors: | Kim, Y, Maltseva, N, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-05 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Crystal Structure of Putative Aldo/Keto Reductase from Salmonella enterica
To be Published
|
|
3PLX
 
 | | The crystal structure of aspartate alpha-decarboxylase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | ACETATE ION, Aspartate 1-decarboxylase, DI(HYDROXYETHYL)ETHER | | Authors: | Tan, K, Gu, M, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-11-15 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | The crystal structure of aspartate alpha-decarboxylase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
4R9X
 
 | | Crystal Structure of Putative Copper Homeostasis Protein CutC from Bacillus anthracis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Copper homeostasis protein CutC, ... | | Authors: | Kim, Y, Zhou, M, Makowska-Grzyska, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-08 | | Release date: | 2014-09-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.8515 Å) | | Cite: | Crystal Structure of Putative Copper Homeostasis Protein CutC
from Bacillus anthracis
To be Published, 2014
|
|
4QVR
 
 | | 2.3 Angstrom Crystal Structure of Hypothetical Protein FTT1539c from Francisella tularensis. | | Descriptor: | Uncharacterized hypothetical protein FTT_1539c | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Flores, K, Ren, G, Huntley, J.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-15 | | Release date: | 2014-07-30 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2.3 Angstrom Crystal Structure of Hypothetical Protein FTT1539c from Francisella tularensis.
TO BE PUBLISHED
|
|
