2Q1M
 
 | |
3MOD
 
 | | Crystal structure of the neutralizing HIV antibody 2F5 Fab fragment (recombinantly produced IgG) with 11 aa gp41 MPER-derived peptide | | Descriptor: | ANTI-HIV-1 ANTIBODY 2F5 HEAVY CHAIN, ANTI-HIV-1 ANTIBODY 2F5 LIGHT CHAIN, gp41 MPER-derived peptide | | Authors: | Nicely, N.I, Dennison, S.M, Kelsoe, G, Liao, H.-X, Alam, S.M, Haynes, B.F. | | Deposit date: | 2010-04-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Non-Neutralizing HIV-1 gp41 Envelope Antibody Demonstrates Neutralization Mechanism of gp41 Antibodies
To be Published
|
|
8T7N
 
 | | Crystal structure of the R132H mutant of IDH1 bound to compound 1 | | Descriptor: | Isocitrate dehydrogenase [NADP] cytoplasmic, N-(4-tert-butylphenyl)-7,8-dimethyl-5,11-dihydro-6H-pyrido[2,3-b][1,5]benzodiazepine-6-carboxamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lu, J, Abeywickrema, P, Heo, M.R, Parthasarathy, G, McCoy, M, Soisson, S.M. | | Deposit date: | 2023-06-20 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Mechanistic and Biostructural Studies of Mutant IDH1
To Be Published
|
|
8T7O
 
 | | Crystal structure of the R132H mutant of IDH1 bound to AG-120 | | Descriptor: | Isocitrate dehydrogenase [NADP] cytoplasmic, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ivosidenib | | Authors: | Lu, J, Abeywickrema, P, Heo, M.R, Parthasarathy, G, McCoy, M, Soisson, S.M. | | Deposit date: | 2023-06-20 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Mechanistic and Biostructural Studies of Mutant IDH1
To Be Published
|
|
8T7D
 
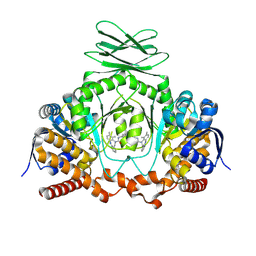 | | Crystal structure of wild type IDH1 bound to compound 1 | | Descriptor: | Isocitrate dehydrogenase [NADP] cytoplasmic, N-(4-tert-butylphenyl)-7,8-dimethyl-5,11-dihydro-6H-pyrido[2,3-b][1,5]benzodiazepine-6-carboxamide | | Authors: | Lu, J, Abeywickrema, P, Heo, M.R, Parthasarathy, G, McCoy, M, Soisson, S.M. | | Deposit date: | 2023-06-20 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (3.444 Å) | | Cite: | Mechanistic and Biostructural Studies of Mutant IDH1
To Be Published
|
|
3MHD
 
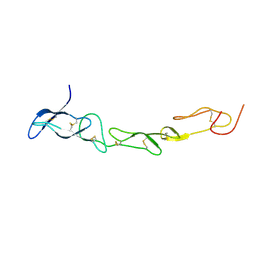 | | Crystal structure of DCR3 | | Descriptor: | Tumor necrosis factor receptor superfamily member 6B | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2010-04-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
3MO1
 
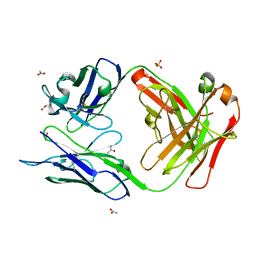 | | Crystal structure of the non-neutralizing HIV antibody 13H11 Fab fragment | | Descriptor: | ACETATE ION, ANTI-HIV-1 ANTIBODY 13H11 HEAVY CHAIN, ANTI-HIV-1 ANTIBODY 13H11 LIGHT CHAIN, ... | | Authors: | Nicely, N.I, Dennison, S.M, Kelsoe, G, Liao, H.-X, Alam, S.M, Haynes, B.F. | | Deposit date: | 2010-04-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Non-Neutralizing HIV-1 gp41 Envelope Antibody Demonstrates Neutralization Mechanism of gp41 Antibodies
To be Published
|
|
2R7S
 
 | | Crystal Structure of Rotavirus SA11 VP1 / RNA (UGUGCC) complex | | Descriptor: | PHOSPHATE ION, RNA (5'-R(*UP*GP*UP*GP*CP*C)-3'), RNA-dependent RNA polymerase | | Authors: | Lu, X, Harrison, S.C, Tao, Y.J, Patton, J.T, Nibert, M.L. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1.
Structure, 16, 2008
|
|
2R7V
 
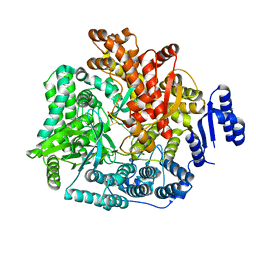 | | Crystal Structure of Rotavirus SA11 VP1/RNA (GGCUUU) Complex | | Descriptor: | RNA (5'-R(*G*GP*CP*UP*UP*U)-3'), RNA-dependent RNA polymerase | | Authors: | Lu, X, Harrison, S.C, Tao, Y.J, Patton, J.T, Nibert, M.L. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1.
Structure, 16, 2008
|
|
4DB5
 
 | | Crystal structure of Rabbit GITRL | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tumor necrosis factor ligand superfamily member 18 | | Authors: | Kumar, P.R, Bhosle, R, Gizzi, A, Scott Glenn, A, Chowhury, S, Hillerich, B, Seidel, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-01-13 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.522 Å) | | Cite: | Crystal structure of GITRL from Oryctolagus cuniculus
to be published
|
|
2VPV
 
 | | Dimerization Domain of Mif2p | | Descriptor: | PROTEIN MIF2, SULFATE ION | | Authors: | Cohen, R.L, Espelin, C.W, Sorger, P.K, Harrison, S.C, Simons, K.T. | | Deposit date: | 2008-03-05 | | Release date: | 2008-08-26 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Dissection of Mif2P, a Conserved DNA-Binding Kinetochore Protein.
Mol.Biol.Cell, 19, 2008
|
|
3MNZ
 
 | | Crystal structure of the non-neutralizing HIV antibody 13H11 Fab fragment with a gp41 MPER-derived peptide bearing Ala substitutions in a helical conformation | | Descriptor: | ANTI-HIV-1 ANTIBODY 13H11 HEAVY CHAIN, ANTI-HIV-1 ANTIBODY 13H11 LIGHT CHAIN, SODIUM ION, ... | | Authors: | Nicely, N.I, Dennison, S.M, Kelsoe, G, Liao, H.-X, Alam, S.M, Haynes, B.F. | | Deposit date: | 2010-04-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Non-Neutralizing HIV-1 gp41 Envelope Antibody Demonstrates Neutralization Mechanism of gp41 Antibodies
To be Published
|
|
2R2A
 
 | | Crystal structure of N-terminal domain of zonular occludens toxin from Neisseria meningitidis | | Descriptor: | SULFATE ION, Uncharacterized protein | | Authors: | Osipiuk, J, Patterson, S, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-24 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of N-terminal domain of zonular occludens toxin from Neisseria meningitidis.
To be Published
|
|
3NWM
 
 | | Crystal structure of a single chain construct composed of MHC class I H-2Kd, beta-2microglobulin and a peptide which is an autoantigen for type 1 diabetes | | Descriptor: | Peptide/beta-2microglobulin/MHC class I H-2Kd chimeric protein | | Authors: | Ramagopal, U.A, Samanta, D, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2010-07-09 | | Release date: | 2011-07-06 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional characterization of a single-chain peptide-MHC molecule that modulates both naive and activated CD8+ T cells.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1TS0
 
 | | Structure of the pB1 intermediate from time-resolved Laue crystallography | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Ihee, H, Rajagopal, S, Srajer, V, Pahl, R, Anderson, S, Schmidt, M, Schotte, F, Anfinrud, P.A, Wulff, M, Moffat, K. | | Deposit date: | 2004-06-21 | | Release date: | 2005-07-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Visualizing reaction pathways in photoactive yellow protein from nanoseconds to seconds.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1TS6
 
 | | Structure of the pB2 intermediate from time-resolved Laue crystallography | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Ihee, H, Rajagopal, S, Srajer, V, Pahl, R, Anderson, S, Schmidt, M, Schotte, F, Anfinrud, P.A, Wulff, M, Moffat, K. | | Deposit date: | 2004-06-21 | | Release date: | 2005-07-05 | | Last modified: | 2021-07-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Visualizing reaction pathways in photoactive yellow protein from nanoseconds to seconds.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1TS8
 
 | | Structure of the pR cis planar intermediate from time-resolved Laue crystallography | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Ihee, H, Rajagopal, S, Srajer, V, Pahl, R, Anderson, S, Schmidt, M, Schotte, F, Anfinrud, P.A, Wulff, M, Moffat, K. | | Deposit date: | 2004-06-21 | | Release date: | 2005-07-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Visualizing reaction pathways in photoactive yellow protein from nanoseconds to seconds.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
7KDF
 
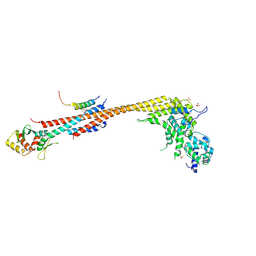 | | Structure of Stu2 Bound to dwarf Ndc80c | | Descriptor: | NDC80 isoform 1,NDC80 isoform 1, NUF2 isoform 1,NUF2 isoform 1, SPC25 isoform 1,SPC25 isoform 1, ... | | Authors: | Zahm, J.A, Stewart, M.G, Miller, M.P, Harrison, S.C. | | Deposit date: | 2020-10-08 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis of Stu2 recruitment to yeast kinetochores.
Elife, 10, 2021
|
|
2YFV
 
 | |
5U0U
 
 | |
5U0R
 
 | |
5TPL
 
 | |
5F6H
 
 | |
5F6J
 
 | |
5UGY
 
 | | Influenza hemagglutinin in complex with a neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH65 heavy chain, ... | | Authors: | Whittle, J.R.R, Jenni, S, Harrison, S.C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin.
Proc. Natl. Acad. Sci. U.S.A., 108, 2011
|
|
