6FFC
 
 | | Structure of an inhibitor-bound ABC transporter | | Descriptor: | ATP-binding cassette sub-family G member 2, ~{tert}-butyl 3-[(2~{S},5~{S},8~{S})-14-cyclopentyloxy-2-(2-methylpropyl)-4,7-bis(oxidanylidene)-3,6,17-triazatetracyclo[8.7.0.0^{3,8}.0^{11,16}]heptadeca-1(10),11,13,15-tetraen-5-yl]propanoate | | Authors: | Jackson, S.M, Manolaridis, I, Kowal, J, Zechner, M, Taylor, N.M.I, Bause, M, Bauer, S, Bartholomaeus, R, Stahlberg, H, Bernhardt, G, Koenig, B, Buschauer, A, Altmann, K.H, Locher, K.P. | | Deposit date: | 2018-01-06 | | Release date: | 2018-04-11 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis of small-molecule inhibition of human multidrug transporter ABCG2.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3IG4
 
 | | Structure of a putative aminopeptidase P from Bacillus anthracis | | Descriptor: | MANGANESE (II) ION, SULFATE ION, Xaa-pro aminopeptidase | | Authors: | Anderson, S.M, Wawrzak, Z, Skarina, T, Onopriyenko, O, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-27 | | Release date: | 2009-08-04 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of a putative aminopeptidase P from Bacillus anthracis
To be Published
|
|
1OTD
 
 | |
4BIK
 
 | | Structure of a disulfide locked mutant of Intermedilysin with human CD59 | | Descriptor: | CD59 GLYCOPROTEIN, INTERMEDILYSIN | | Authors: | Johnson, S, Brooks, N.J, Smith, R.A.G, Lea, S.M, Bubeck, D. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.494 Å) | | Cite: | Structural Basis for Recognition of the Pore- Forming Toxin Intermedilysin by Human Complement Receptor Cd59
Cell Rep., 3, 2013
|
|
4O96
 
 | | 2.60 Angstrom resolution crystal structure of a protein kinase domain of type III effector NleH2 (ECs1814) from Escherichia coli O157:H7 str. Sakai | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, type III effector protein kinase | | Authors: | Anderson, S.M, Halavaty, A.S, Wawrzak, Z, Kudritska, M, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-01 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Type III Effector NleH2 from Escherichia coli O157:H7 str. Sakai Features an Atypical Protein Kinase Domain.
Biochemistry, 53, 2014
|
|
7UPM
 
 | |
7UWL
 
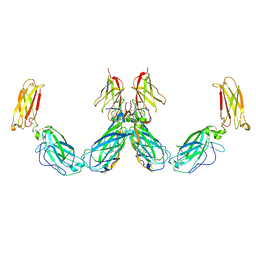 | | Structure of the IL-25-IL-17RB-IL-17RA ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor A, ... | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
3HZN
 
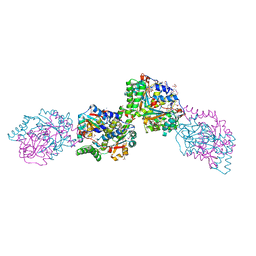 | | Structure of the Salmonella typhimurium nfnB dihydropteridine reductase | | Descriptor: | ACETATE ION, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Skarina, T, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Salmonella typhimurium nfnB dihydropteridine reductase
TO BE PUBLISHED
|
|
5FHK
 
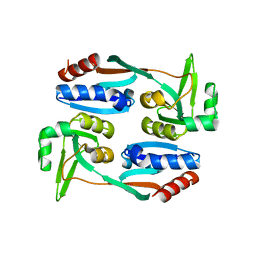 | | Regulatory domain of AphB in Vibrio vulnificus | | Descriptor: | LysR family transcriptional regulator | | Authors: | Song, S, Ha, N.-C. | | Deposit date: | 2015-12-22 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Crystal Structure of the Regulatory Domain of AphB from Vibrio vulnificus, a Virulence Gene Regulator
Mol. Cells, 40, 2017
|
|
3N5M
 
 | | Crystals structure of a Bacillus anthracis aminotransferase | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, CHLORIDE ION, SULFATE ION | | Authors: | Anderson, S.M, Wawrzak, Z, DiLeo, R, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-25 | | Release date: | 2010-06-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystals structure of a Bacillus anthracis aminotransferase
TO BE PUBLISHED
|
|
6F2D
 
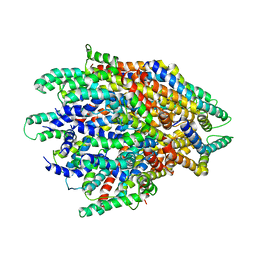 | | A FliPQR complex forms the core of the Salmonella type III secretion system export apparatus. | | Descriptor: | Flagellar biosynthetic protein FliP, Flagellar biosynthetic protein FliQ, Flagellar biosynthetic protein FliR | | Authors: | Johnson, S, Kuhlen, L, Abrusci, P, Lea, S.M. | | Deposit date: | 2017-11-24 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the core of the type III secretion system export apparatus.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6F9B
 
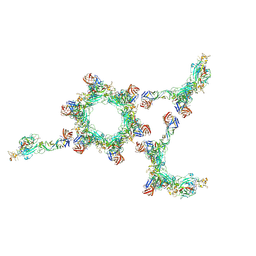 | |
6F9E
 
 | |
6F9D
 
 | |
3Q7H
 
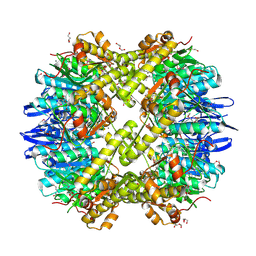 | | Structure of the ClpP subunit of the ATP-dependent Clp Protease from Coxiella burnetii | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CALCIUM ION, DI(HYDROXYETHYL)ETHER | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Hasseman, J, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the ClpP subunit of the ATP-dependent Clp Protease from Coxiella burnetii
To be Published
|
|
6F8P
 
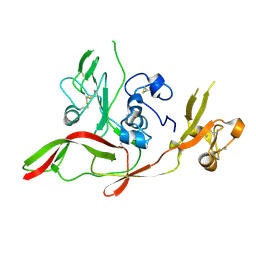 | |
6F9C
 
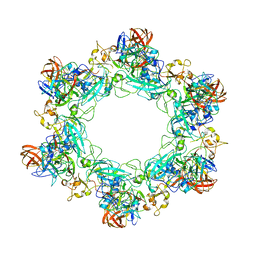 | |
6F9F
 
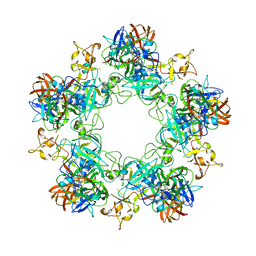 | |
6FEQ
 
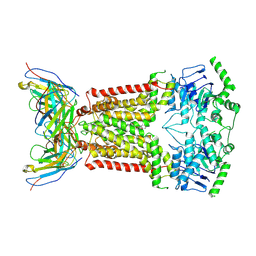 | | Structure of inhibitor-bound ABCG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3(Fab) heavy chain variable domain, 5D3(Fab) light chain variable domain, ... | | Authors: | Jackson, S.M, Manolaridis, I, Kowal, J, Zechner, M, Altmann, K.H, Locher, K.P. | | Deposit date: | 2018-01-03 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of small-molecule inhibition of human multidrug transporter ABCG2.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3OSU
 
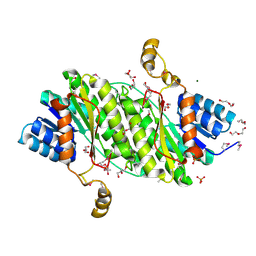 | | Crystal structure of the 3-oxoacyl-acyl carrier protein reductase, FabG, from Staphylococcus aureus | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Edwards, A, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-09 | | Release date: | 2010-09-29 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the 3-oxoacyl-acyl carrier protein reductase, FabG, from Staphylococcus aureus
To be Published
|
|
3OJC
 
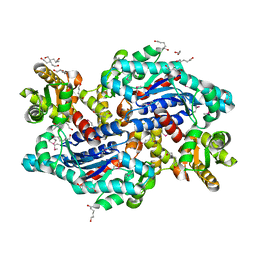 | | Crystal structure of a putative Asp/Glu Racemase from Yersinia pestis | | Descriptor: | CALCIUM ION, HEXANE-1,6-DIOL, Putative aspartate/glutamate racemase | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-21 | | Release date: | 2010-09-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a putative Asp/Glu Racemase from Yersinia pestis
To be Published
|
|
3O04
 
 | | Crystal structure of the beta-keto-acyl carrier protein synthase II (lmo2201) from Listeria monocytogenes | | Descriptor: | beta-keto-acyl carrier protein synthase II | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-19 | | Release date: | 2010-08-04 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the beta-keto-acyl carrier protein synthase II (lmo2201) from Listeria monocytogenes
To be Published
|
|
7UWM
 
 | | Structure of the IL-17A-IL-17RA binary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor A, ... | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
7UWK
 
 | | Structure of the higher-order IL-25-IL-17RB complex | | Descriptor: | Interleukin-17 receptor B, Interleukin-25 | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
7UWJ
 
 | | Structure of the homodimeric IL-25-IL-17RB binary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor B, Interleukin-25 | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
