1QQG
 
 | |
1KS5
 
 | | Structure of Aspergillus niger endoglucanase | | Descriptor: | Endoglucanase A | | Authors: | Khademi, S, Zhang, D, Swanson, S.M, Wartenberg, A, Witte, C, Meyer, E.F. | | Deposit date: | 2002-01-10 | | Release date: | 2003-01-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Determination of the structure of an endoglucanase from Aspergillus niger and its mode of inhibition by palladium chloride.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KS4
 
 | | The structure of Aspergillus niger endoglucanase-palladium complex | | Descriptor: | Endoglucanase A, PALLADIUM ION | | Authors: | Khademi, S, Zhang, D, Swanson, S.M, Wartenberg, A, Witte, C, Meyer, E.F. | | Deposit date: | 2002-01-10 | | Release date: | 2003-01-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determination of the structure of an endoglucanase from Aspergillus niger and its mode of inhibition by palladium chloride.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5UG0
 
 | | Human antibody H2897 in complex with influenza hemagglutinin H1 Solomon Islands/03/2006 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2897 heavy chain, ... | | Authors: | Raymond, D.D, Caradonna, T, Schmidt, A.G, Harrison, S.C. | | Deposit date: | 2017-01-06 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
3T0Y
 
 | |
3OXH
 
 | | Mycobacterium tuberculosis kinase inhibitor homolog RV0577 | | Descriptor: | CHLORIDE ION, PARA-MERCURY-BENZENESULFONIC ACID, RV0577 PROTEIN, ... | | Authors: | Echols, N, Flynn, E.M, Stephenson, S, Ng, H.-L, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-09-21 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Biophysical Characterization of the Mycobacterium tuberculosis Protein Rv0577, a Protein Associated with Neutral Red Staining of Virulent Tuberculosis Strains and Homologue of the Streptomyces coelicolor Protein KbpA.
Biochemistry, 2017
|
|
7LFH
 
 | |
1DZL
 
 | |
4FHQ
 
 | | Crystal Structure of HVEM | | Descriptor: | Tumor necrosis factor receptor superfamily member 14 | | Authors: | Liu, W, Zhan, C, Patskovsky, Y, Bhosle, R.C, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-06-06 | | Release date: | 2012-07-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Increased Heterologous Protein Expression in Drosophila S2 Cells for Massive Production of Immune Ligands/Receptors and Structural Analysis of Human HVEM.
Mol Biotechnol, 57, 2015
|
|
8JU4
 
 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 1,4-androstadiene-3,17- dione | | Descriptor: | 3-ketosteroid dehydrogenase, ANDROSTA-1,4-DIENE-3,17-DIONE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hu, Y.L, Li, X, Cheng, X.Y, Song, S.K, Su, Z.D. | | Deposit date: | 2023-06-24 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 1,4-androstadiene-3,17- dione
To Be Published
|
|
1RE1
 
 | |
6FO6
 
 | | CryoEM structure of bovine cytochrome bc1 in complex with the anti-malarial inhibitor SCR0911 | | Descriptor: | 7-methoxy-3-methyl-2-[5-[4-(trifluoromethyloxy)phenyl]pyridin-3-yl]quinolin-4-ol, Cytochrome b, Cytochrome b-c1 complex subunit 1, ... | | Authors: | Johnson, R.M, Amporndanai, K, O'Neill, P.M, Fishwick, C.W.G, Jamson, A.H, Rawson, S.D, Hasnain, S.S, Antonyuk, S.V, Muench, S.P. | | Deposit date: | 2018-02-06 | | Release date: | 2018-02-28 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | X-ray and cryo-EM structures of inhibitor-bound cytochromebc1complexes for structure-based drug discovery.
IUCrJ, 5, 2018
|
|
1R69
 
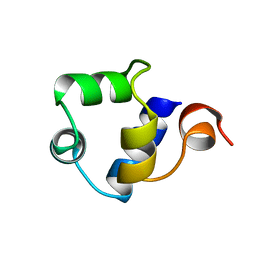 | | STRUCTURE OF THE AMINO-TERMINAL DOMAIN OF PHAGE 434 REPRESSOR AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | REPRESSOR PROTEIN CI | | Authors: | Mondragon, A, Subbiah, S, Alamo, S.C, Drottar, M, Harrison, S.C. | | Deposit date: | 1988-12-08 | | Release date: | 1989-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the amino-terminal domain of phage 434 repressor at 2.0 A resolution.
J.Mol.Biol., 205, 1989
|
|
4G4F
 
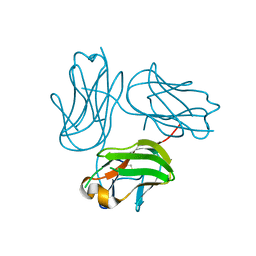 | |
4AVK
 
 | | Structure of trigonal FimH lectin domain crystal soaked with an alpha- D-mannoside O-linked to propynyl pyridine at 2.4A resolution | | Descriptor: | 3-(pyridin-3-yl)prop-2-yn-1-yl alpha-D-mannopyranoside, FIMH, NICKEL (II) ION, ... | | Authors: | Wellens, A, Lahmann, M, Touaibia, M, Vaucher, J, Oscarson, S, Roy, R, Remaut, H, Bouckaert, J. | | Deposit date: | 2012-05-26 | | Release date: | 2012-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Tyrosine Gate as a Potential Entropic Lever in the Receptor-Binding Site of the Bacterial Adhesin Fimh.
Biochemistry, 51, 2012
|
|
4IT3
 
 | | Crystal Structure of Iml3 from S. cerevisiae | | Descriptor: | Central kinetochore subunit IML3 | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2013-01-17 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | An iml3-chl4 heterodimer links the core centromere to factors required for accurate chromosome segregation.
Cell Rep, 5, 2013
|
|
6FO0
 
 | | CryoEM structure of bovine cytochrome bc1 in complex with the anti-malarial compound GSK932121 | | Descriptor: | 3-chloro-6-(hydroxymethyl)-2-methyl-5-{4-[3-(trifluoromethoxy)phenoxy]phenyl}pyridin-4-ol, Chain I/V, Cytochrome b, ... | | Authors: | Johnson, R.M, Amporndanai, K, O'Neill, P.M, Fishwick, C.W.G, Jamson, A.H, Rawson, S.D, Hasnain, S.S, Antonyuk, S.V, Muench, S.P. | | Deposit date: | 2018-02-05 | | Release date: | 2018-02-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | X-ray and cryo-EM structures of inhibitor-bound cytochromebc1complexes for structure-based drug discovery.
IUCrJ, 5, 2018
|
|
6FO2
 
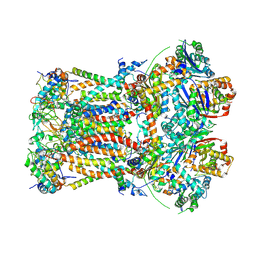 | | CryoEM structure of bovine cytochrome bc1 with no ligand bound | | Descriptor: | Cytochrome b, Cytochrome b-c1 complex subunit 1, mitochondrial, ... | | Authors: | Johnson, R.M, Amporndanai, K, O'Neill, P.M, Fishwick, C.W.G, Jamson, A.H, Rawson, S.D, Hasnain, S.S, Antonyuk, S.V, Muench, S.P. | | Deposit date: | 2018-02-05 | | Release date: | 2018-02-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | X-ray and cryo-EM structures of inhibitor-bound cytochromebc1complexes for structure-based drug discovery.
IUCrJ, 5, 2018
|
|
4B8E
 
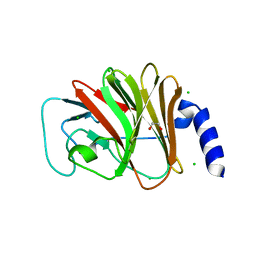 | | PRY-SPRY domain of Trim25 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, E3 UBIQUITIN/ISG15 LIGASE TRIM25 | | Authors: | Kershaw, N.J, D'Cruz, A.A, Nicola, N.A, Nicholson, S.E, Babon, J.J. | | Deposit date: | 2012-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Crystal Structure of the Trim25 B30.2 (Pryspry) Domain: A Key Component of Antiviral Signaling.
Biochem.J., 456, 2013
|
|
1RHJ
 
 | |
5DKT
 
 | | N-terminal His tagged apPOL exonuclease mutant | | Descriptor: | Prex DNA polymerase, SODIUM ION, SULFATE ION | | Authors: | Milton, M.E, Honzatko, R.B, Choe, J.Y, Nelson, S.W. | | Deposit date: | 2015-09-04 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Apicoplast DNA Polymerase from Plasmodium falciparum: The First Look at a Plastidic A-Family DNA Polymerase.
J.Mol.Biol., 428, 2016
|
|
4NEW
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma cruzi in complex with inhibitor EP127 (5-{5-[1-(PYRROLIDIN-1-YL)CYCLOHEXYL]-1,3-THIAZOL-2-YL}-1H-INDOLE) | | Descriptor: | 5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole, FLAVIN-ADENINE DINUCLEOTIDE, Trypanothione reductase, ... | | Authors: | Persch, E, Bryson, S, Pai, E.F, Krauth-Siegel, R.L, Diederich, F. | | Deposit date: | 2013-10-30 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding to large enzyme pockets: small-molecule inhibitors of trypanothione reductase.
Chemmedchem, 9, 2014
|
|
2C1N
 
 | | Molecular basis for the recognition of phosphorylated and phosphoacetylated histone H3 by 14-3-3 | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, HISTONE H3 ACETYLPHOSPHOPEPTIDE | | Authors: | Welburn, J.P.I, Macdonald, N, Noble, M.E.M, Nguyen, A, Yaffe, M.B, Clynes, D, Moggs, J.G, Orphanides, G, Thomson, S, Edmunds, J.W, Clayton, A.L, Endicott, J.A, Mahadevan, L.C. | | Deposit date: | 2005-09-16 | | Release date: | 2005-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Basis for the Recognition of Phosphorylated and Phosphoacetylated Histone H3 by 14-3-3.
Mol.Cell, 20, 2005
|
|
4AVI
 
 | | Structure of the FimH lectin domain in the trigonal space group, in complex with a methyl ester octyl alpha-D-mannoside at 2.4 A resolution | | Descriptor: | FIMH, METHYL ESTER OCTYL ALPHA-1O-D-MANNOPYRANNOSIDE, NICKEL (II) ION, ... | | Authors: | Wellens, A, Lahmann, M, Touaibia, M, Vaucher, J, Oscarson, S, Roy, R, Remaut, H, Bouckaert, J. | | Deposit date: | 2012-05-26 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Tyrosine Gate as a Potential Entropic Lever in the Receptor-Binding Site of the Bacterial Adhesin Fimh.
Biochemistry, 51, 2012
|
|
2C1J
 
 | | Molecular basis for the recognition of phosphorylated and phosphoacetylated histone H3 by 14-3-3 | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, HISTONE H3 ACETYLPHOSPHOPEPTIDE | | Authors: | Welburn, J.P.I, Macdonald, N, Noble, M.E.M, Nguyen, A, Yaffe, M.B, Clynes, D, Moggs, J.G, Orphanides, G, Thomson, S, Edmunds, J.W, Clayton, A.L, Endicott, J.A, Mahadevan, L.C. | | Deposit date: | 2005-09-15 | | Release date: | 2005-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Basis for the Recognition of Phosphorylated and Phosphoacetylated Histone H3 by 14-3-3.
Mol.Cell, 20, 2005
|
|
