3MP9
 
 | | Structure of Streptococcal protein G B1 domain at pH 3.0 | | Descriptor: | FORMIC ACID, Immunoglobulin G-binding protein G | | Authors: | Tomlinson, J.H, Green, V.L, Baker, P.J, Williamson, M.P. | | Deposit date: | 2010-04-26 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural origins of pH-dependent chemical shifts in the B1 domain of protein G.
Proteins, 78, 2010
|
|
5IZ2
 
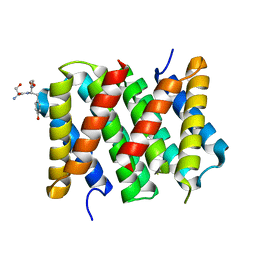 | | Crystal structure of the N. clavipes spidroin NTD at pH 6.5 | | Descriptor: | Major ampullate spidroin 1A, Major ampullate spidroin 1A (Partial C-terminus) | | Authors: | Atkison, J.H, Olsen, S.K. | | Deposit date: | 2016-03-24 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structure of the Nephila clavipes Major Ampullate Spidroin 1A N-terminal Domain Reveals Plasticity at the Dimer Interface.
J.Biol.Chem., 291, 2016
|
|
2MZW
 
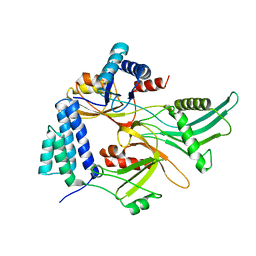 | | Staphylococcus aureus FusB:EF-GC3 complex | | Descriptor: | Elongation factor G, Far1, ZINC ION | | Authors: | Tomlinson, J.H, Thompson, G.S, Kalverda, A.P, Zhuravleva, A, O'Neill, A. | | Deposit date: | 2015-02-25 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A target-protection mechanism of antibiotic resistance at atomic resolution: insights into FusB-type fusidic acid resistance.
Sci Rep, 6, 2016
|
|
1UC7
 
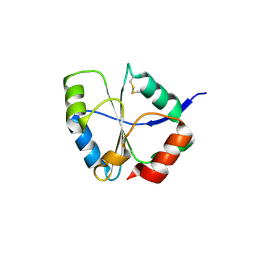 | | Crystal structure of DsbDgamma | | Descriptor: | Thiol:disulfide interchange protein dsbD | | Authors: | Kim, J.H, Kim, S.J, Jeong, D.G, Son, J.H, Ryu, S.E. | | Deposit date: | 2003-04-09 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of DsbDgamma reveals the mechanism of redox potential shift and substrate specificity(1)
FEBS LETT., 543, 2003
|
|
4K86
 
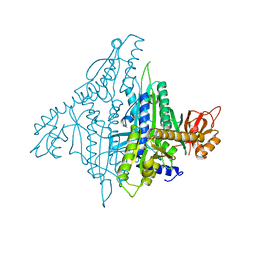 | |
4K87
 
 | | Crystal structure of human prolyl-tRNA synthetase (substrate bound form) | | Descriptor: | ADENOSINE, PROLINE, Proline--tRNA ligase, ... | | Authors: | Hwang, K.Y, Son, J.H, Lee, E.H. | | Deposit date: | 2013-04-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Conformational changes in human prolyl-tRNA synthetase upon binding of the substrates proline and ATP and the inhibitor halofuginone.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4K88
 
 | | Crystal structure of human prolyl-tRNA synthetase (halofuginone bound form) | | Descriptor: | 7-bromo-6-chloro-3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, Proline--tRNA ligase, ZINC ION | | Authors: | Hwang, K.Y, Son, J.H, Lee, E.H. | | Deposit date: | 2013-04-18 | | Release date: | 2013-10-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.619 Å) | | Cite: | Conformational changes in human prolyl-tRNA synthetase upon binding of the substrates proline and ATP and the inhibitor halofuginone.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1XM2
 
 | | Crystal structure of Human PRL-1 | | Descriptor: | SULFATE ION, Tyrosine Phosphatase | | Authors: | Jeong, D.G, Kim, S.J, Kim, J.H, Son, J.H, Ryu, S.E. | | Deposit date: | 2004-10-01 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Trimeric structure of PRL-1 phosphatase reveals an active enzyme conformation and regulation mechanisms
J.Mol.Biol., 345, 2005
|
|
1ZZW
 
 | | Crystal Structure of catalytic domain of Human MAP Kinase Phosphatase 5 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein phosphatase 10, SULFATE ION | | Authors: | Jeong, D.G, Yoon, T.S, Kim, J.H, Shim, M.Y, Jeong, S.K, Son, J.H, Ryu, S.E, Kim, S.J. | | Deposit date: | 2005-06-14 | | Release date: | 2006-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human MAP Kinase Phosphatase 5: Structural Insight into Constitutively Active Phosphatase.
J.Mol.Biol., 360, 2006
|
|
1YZ4
 
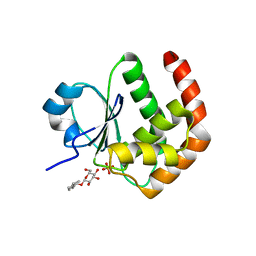 | | Crystal structure of DUSP15 | | Descriptor: | SULFATE ION, dual specificity phosphatase-like 15 isoform a, octyl beta-D-glucopyranoside | | Authors: | Kim, S.J, Ryu, S.E, Jeong, D.G, Yoon, T.S, Kim, J.H, Cho, Y.H, Jeong, S.K, Lee, J.W, Son, J.H. | | Deposit date: | 2005-02-28 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the catalytic domain of human VHY, a dual-specificity protein phosphatase
Proteins, 61, 2005
|
|
5KXA
 
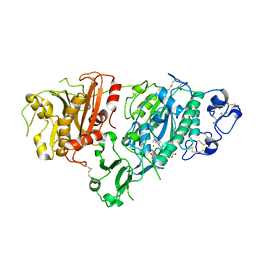 | | Selective Inhibition of Autotaxin is Effective in Mouse Models of Liver Fibrosis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[6-chloranyl-2-cyclopropyl-1-(1-ethylpyrazol-4-yl)-7-fluoranyl-indol-3-yl]sulfanyl-2-fluoranyl-benzoic acid, CALCIUM ION, ... | | Authors: | Stein, A.J, Bain, G, Hutchinson, J.H, Evans, J.F. | | Deposit date: | 2016-07-20 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Selective Inhibition of Autotaxin Is Efficacious in Mouse Models of Liver Fibrosis.
J. Pharmacol. Exp. Ther., 360, 2017
|
|
4ZGA
 
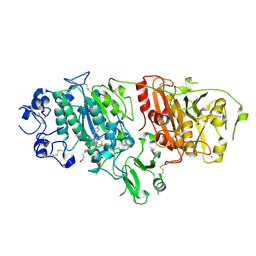 | | Structural basis for inhibition of human autotaxin by four novel compounds | | Descriptor: | (11aS)-6-(4-fluorobenzyl)-5,6,11,11a-tetrahydro-1H-imidazo[1',5':1,6]pyrido[3,4-b]indole-1,3(2H)-dione, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stein, A.J, Bain, G, Hutchinson, J.H, Evans, J.F. | | Deposit date: | 2015-04-22 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Inhibition of Human Autotaxin by Four Potent Compounds with Distinct Modes of Binding.
Mol.Pharmacol., 88, 2015
|
|
4ZG7
 
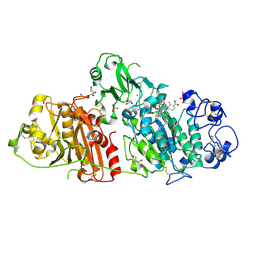 | | Structural basis for inhibition of human autotaxin by four novel compounds | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-({6-chloro-7-fluoro-2-methyl-1-[2-oxo-2-(spiro[cyclopropane-1,3'-indol]-1'(2'H)-yl)ethyl]-1H-indol-3-yl}sulfanyl)-2-fluorobenzoic acid, ... | | Authors: | Stein, A.J, Bain, G, Hutchinson, J.H, Evans, J.F. | | Deposit date: | 2015-04-22 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Inhibition of Human Autotaxin by Four Potent Compounds with Distinct Modes of Binding.
Mol.Pharmacol., 88, 2015
|
|
4ZG6
 
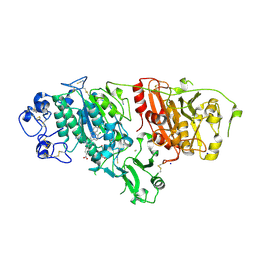 | | Structural basis for inhibition of human autotaxin by four novel compounds | | Descriptor: | 4-{(Z)-2-[6-chloro-1-(4-fluorobenzyl)-1H-indol-3-yl]-1-cyanoethenyl}benzoic acid, CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Stein, A.J, Bain, G, Hutchinson, J.H, Evans, J.F. | | Deposit date: | 2015-04-22 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Inhibition of Human Autotaxin by Four Potent Compounds with Distinct Modes of Binding.
Mol.Pharmacol., 88, 2015
|
|
4ZG9
 
 | | Structural basis for inhibition of human autotaxin by four novel compounds | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(11aS)-6-(4-fluorobenzyl)-1,3-dioxo-5,6,11,11a-tetrahydro-1H-imidazo[1',5':1,6]pyrido[3,4-b]indol-2(3H)-yl]propanoic acid, ... | | Authors: | Stein, A.J, Bain, G, Hutchinson, J.H, Evans, J.F. | | Deposit date: | 2015-04-22 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Basis for Inhibition of Human Autotaxin by Four Potent Compounds with Distinct Modes of Binding.
Mol.Pharmacol., 88, 2015
|
|
4LKM
 
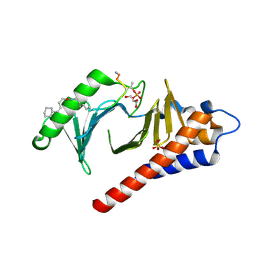 | | Crystal structure of Plk1 Polo-box domain in complex with PL-74 | | Descriptor: | GLYCEROL, PL-74, SULFATE ION, ... | | Authors: | Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2013-07-08 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the binding nature of pyrrolidine pocket-dependent interactions in the polo-box domain of polo-like kinase 1
Plos One, 8, 2013
|
|
4LKL
 
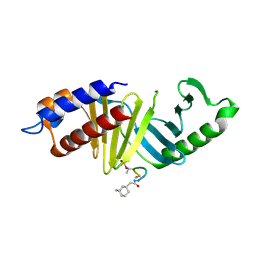 | |
5IDH
 
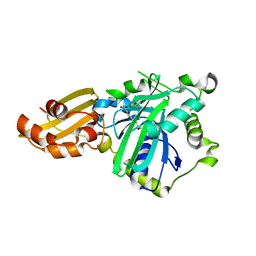 | |
5ICL
 
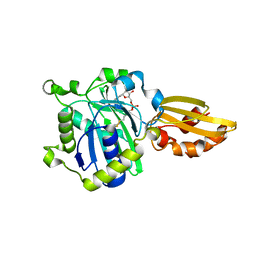 | |
5ICH
 
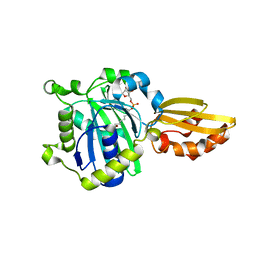 | |
6NYA
 
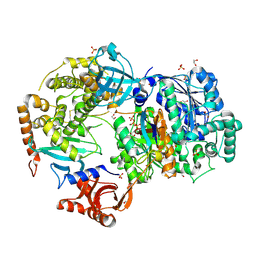 | | Crystal Structure of ubiquitin E1 (Uba1) in complex with Ubc3 (Cdc34) and ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Olsen, S.K, Williams, K.M, Atkison, J.H. | | Deposit date: | 2019-02-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.065 Å) | | Cite: | Structural insights into E1 recognition and the ubiquitin-conjugating activity of the E2 enzyme Cdc34.
Nat Commun, 10, 2019
|
|
6NYO
 
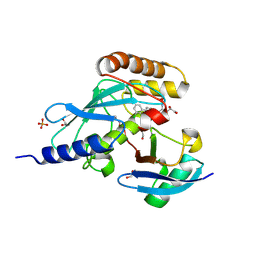 | | Crystal structure of a human Cdc34-ubiquitin thioester mimetic | | Descriptor: | 1,2-ETHANEDIOL, 4,5-dideoxy-5-(3',5'-dichlorobiphenyl-4-yl)-4-[(methoxyacetyl)amino]-L-arabinonic acid, PHOSPHATE ION, ... | | Authors: | Olsen, S.K, Williams, K.M, Atkison, J.H. | | Deposit date: | 2019-02-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Structural insights into E1 recognition and the ubiquitin-conjugating activity of the E2 enzyme Cdc34.
Nat Commun, 10, 2019
|
|
5IBY
 
 | |
6NYD
 
 | |
6CWY
 
 | | Crystal structure of SUMO E1 in complex with an allosteric inhibitor | | Descriptor: | GLYCEROL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Lv, Z, Yuan, L, Atkison, J.H, Williams, K.M, Olsen, S.K. | | Deposit date: | 2018-04-01 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.462 Å) | | Cite: | Molecular mechanism of a covalent allosteric inhibitor of SUMO E1 activating enzyme.
Nat Commun, 9, 2018
|
|
