2GDM
 
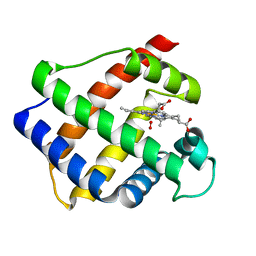 | | LEGHEMOGLOBIN (OXY) | | Descriptor: | LEGHEMOGLOBIN (OXY), OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Harutyunyan, E.H, Safonova, T.N, Kuranova, I.P, Popov, A.N, Teplyakov, A.V, Obmolova, G.V, Rusakov, A.A, Dodson, G.G, Wilson, J.C, Perutz, M.F. | | Deposit date: | 1994-09-14 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of Deoxy-and Oxy-Leghaemoglobin from Lupin
J.Mol.Biol., 251, 1995
|
|
2RGJ
 
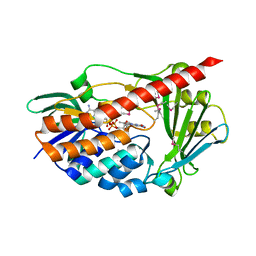 | | Crystal structure of flavin-containing monooxygenase PhzS | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase | | Authors: | Ladner, J.E, Parsons, J.F, Greenhagen, B.T, Robinson, H. | | Deposit date: | 2007-10-03 | | Release date: | 2008-05-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Pyocyanin Biosynthetic Protein PhzS.
Biochemistry, 47, 2008
|
|
4K86
 
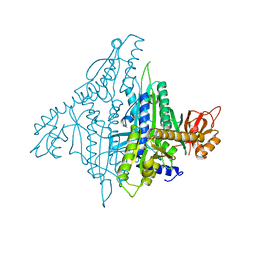 | |
4K88
 
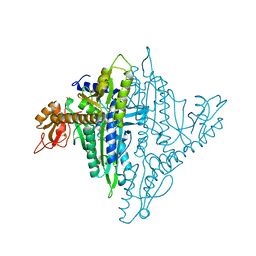 | | Crystal structure of human prolyl-tRNA synthetase (halofuginone bound form) | | Descriptor: | 7-bromo-6-chloro-3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, Proline--tRNA ligase, ZINC ION | | Authors: | Hwang, K.Y, Son, J.H, Lee, E.H. | | Deposit date: | 2013-04-18 | | Release date: | 2013-10-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.619 Å) | | Cite: | Conformational changes in human prolyl-tRNA synthetase upon binding of the substrates proline and ATP and the inhibitor halofuginone.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4K87
 
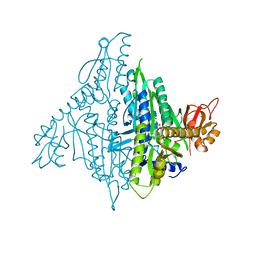 | | Crystal structure of human prolyl-tRNA synthetase (substrate bound form) | | Descriptor: | ADENOSINE, PROLINE, Proline--tRNA ligase, ... | | Authors: | Hwang, K.Y, Son, J.H, Lee, E.H. | | Deposit date: | 2013-04-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Conformational changes in human prolyl-tRNA synthetase upon binding of the substrates proline and ATP and the inhibitor halofuginone.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1MPL
 
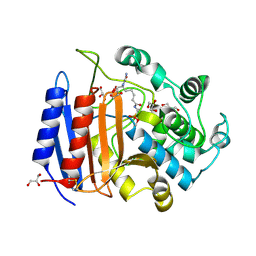 | | CRYSTAL STRUCTURE OF PHOSPHONATE-INHIBITED D-ALA-D-ALA PEPTIDASE REVEALS AN ANALOG OF A TETRAHEDRAL TRANSITION STATE | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, GLYCEROL, GLYCYL-L-A-AMINOPIMELYL-E-(D-2-AMINOETHYL)PHOSPHONATE | | Authors: | Silvaggi, N.R, Anderson, J.W, Brinsmade, S.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2002-09-12 | | Release date: | 2003-02-25 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | The Crystal Structure of Phosphonate-Inhibited
d-Ala-d-Ala Peptidase Reveals an Analogue of a Tetrahedral Transition State.
Biochemistry, 42, 2003
|
|
5W0V
 
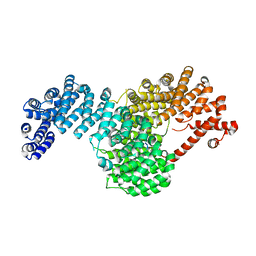 | | Crystal structure of full-length Kluyveromyces lactis Kap123 with histone H4 1-34 | | Descriptor: | Histone H4 1-34, Kap123 | | Authors: | An, S, Yoon, J, Song, J.-J, Cho, U.-S. | | Deposit date: | 2017-05-31 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.821 Å) | | Cite: | Structure-based nuclear import mechanism of histones H3 and H4 mediated by Kap123.
Elife, 6, 2017
|
|
6R8I
 
 | | PP4R3A EVH1 domain bound to FxxP motif | | Descriptor: | SER-LEU-PRO-PHE-THR-PHE-LYS-VAL-PRO-ALA-PRO-PRO-PRO-SER-LEU-PRO-PRO-SER, Serine/threonine-protein phosphatase 4 regulatory subunit 3A | | Authors: | Ueki, Y, Kruse, T, Weisser, M.B, Sundell, G.N, Yoo Larsen, M.S, Lopez Mendez, B, Jenkins, N.P, Garvanska, D.H, Cressey, L, Zhang, G, Davey, N, Montoya, G, Ivarsson, Y, Kettenbach, A, Nilsson, J. | | Deposit date: | 2019-04-02 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.517 Å) | | Cite: | A Consensus Binding Motif for the PP4 Protein Phosphatase.
Mol.Cell, 76, 2019
|
|
5AG4
 
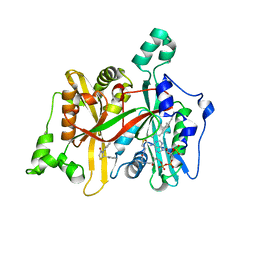 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A THIAZOLIDINONE LIGAND | | Descriptor: | (2S)-3-(3-chlorophenyl)-2-(pyridin-2-yl)-1,3-thiazolidin-4-one, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Spinks, D, Smith, V.C, Thompson, S, Smith, A, Torrie, L.S, McElroy, S.P, Brand, S, Brenk, R, Frearson, J.A, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2015-01-29 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Development of Small-Molecule Trypanosoma Brucei N-Myristoyltransferase Inhibitors: Discovery and Optimisation of a Novel Binding Mode.
Chemmedchem, 10, 2015
|
|
5AGE
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A BENZOMORPHOLINONE LIGAND | | Descriptor: | 4-[(5-methyl-1,2-oxazol-3-yl)methyl]-7-[4-(1-methylpiperidin-4-yl)butyl]-2H-1,4-benzoxazin-3(4H)-one, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Spinks, D, Smith, V.C, Thompson, S, Smith, A, Torrie, L.S, McElroy, S.P, Brand, S, Brenk, R, Frearson, J.A, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2015-01-29 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of Small-Molecule Trypanosoma Brucei N-Myristoyltransferase Inhibitors: Discovery and Optimisation of a Novel Binding Mode.
Chemmedchem, 10, 2015
|
|
5AG7
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A BENZOMORPHOLINE LIGAND | | Descriptor: | GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA, ethyl (3-oxo-2,3-dihydro-4H-1,4-benzoxazin-4-yl)acetate | | Authors: | Robinson, D.A, Spinks, D, Smith, V.C, Thompson, S, Smith, A, Torrie, L.S, McElroy, S.P, Brand, S, Brenk, R, Frearson, J.A, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2015-01-29 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development of Small-Molecule Trypanosoma Brucei N-Myristoyltransferase Inhibitors: Discovery and Optimisation of a Novel Binding Mode.
Chemmedchem, 10, 2015
|
|
1W8N
 
 | | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens. | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, BACTERIAL SIALIDASE, SODIUM ION, ... | | Authors: | Newstead, S, Watson, J.N, Dookhun, V, Bennet, A.J, Taylor, G. | | Deposit date: | 2004-09-24 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora Viridifaciens
FEBS Lett., 577, 2004
|
|
1N1L
 
 | | CRYSTAL STRUCTURE OF HCV NS3 PROTEASE DOMAIN:NS4A PEPTIDE COMPLEX WITH COVALENTLY BOUND INHIBITOR (GW472467X) | | Descriptor: | HCV NS3 SERINE PROTEASE, NS4A COFACTOR, ZINC ION, ... | | Authors: | Andrews, D.M, Chaignot, H, Coomber, B.A, Good, A.C, Hind, S.L, Jones, P.S, Mill, G, Robinson, J.E, Skarzynski, T, Slater, M.J, Somers, D.O.N. | | Deposit date: | 2002-10-18 | | Release date: | 2003-10-21 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pyrrolidine-5,5-trans-lactams. 2. The use of X-ray Crystal Structure Data in the Optimisation of P3 and P4 Substituents
Org.Lett., 4, 2002
|
|
5W89
 
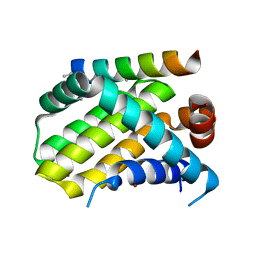 | | Crystal structure of human Mcl-1 in complex with modified Bim BH3 peptide SAH-MS1-18 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION, modified Bim BH3 peptide SAH-MS1-18 | | Authors: | Rezaei Araghi, R, Jenson, J.M, Grant, R.A, Keating, A.E. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Iterative optimization yields Mcl-1-targeting stapled peptides with selective cytotoxicity to Mcl-1-dependent cancer cells.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5W8F
 
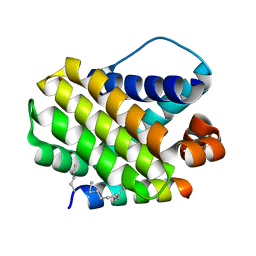 | | Crystal structure of human Mcl-1 in complex with modified Bim BH3 peptide SAH-MS1-14 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION, modified Bim BH3 peptide SAH-MS1-14 | | Authors: | Rezaei Araghi, R, Jenson, J.M, Grant, R.A, Keating, A.E. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Iterative optimization yields Mcl-1-targeting stapled peptides with selective cytotoxicity to Mcl-1-dependent cancer cells.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5FA9
 
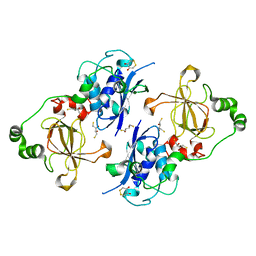 | | Bifunctional Methionine Sulfoxide Reductase AB (MsrAB) from Treponema denticola | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Peptide methionine sulfoxide reductase MsrA | | Authors: | Han, A, Son, J, Kim, H.-Y, Hwang, K.Y. | | Deposit date: | 2015-12-11 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Essential Role of the Linker Region in the Higher Catalytic Efficiency of a Bifunctional MsrA-MsrB Fusion Protein
Biochemistry, 55, 2016
|
|
1EMZ
 
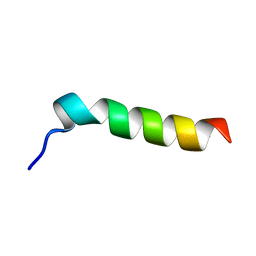 | | SOLUTION STRUCTURE OF FRAGMENT (350-370) OF THE TRANSMEMBRANE DOMAIN OF HEPATITIS C ENVELOPE GLYCOPROTEIN E1 | | Descriptor: | ENVELOPE GLYCOPROTEIN E1 | | Authors: | Op De Beeck, A, Montserret, R, Duvet, S, Cocquerel, L, Cacan, R, Barberot, B, Le Maire, M, Penin, F, Dubuisson, J. | | Deposit date: | 2000-03-20 | | Release date: | 2000-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The transmembrane domains of hepatitis C virus envelope glycoproteins E1 and E2 play a major role in heterodimerization.
J.Biol.Chem., 275, 2000
|
|
6DN1
 
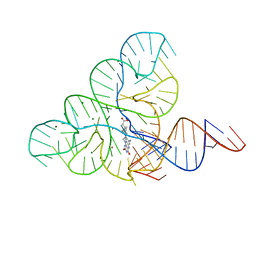 | | CRYSTAL STRUCTURE OF THE FMN RIBOSWITCH BOUND TO BRX1151 SPLIT RNA | | Descriptor: | 10-(6-carboxyhexyl)-8-(cyclopentylamino)-2,4-dihydroxy-7-methylbenzo[g]pteridin-10-ium, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Vicens, Q, Mondragon, E, Reyes, F.E, Berman, J, Kaur, H, Kells, K, Wickens, P, Wilson, J, Gadwood, R, Schostarez, H, Suto, R.K, Coish, P, Blount, K.F, Batey, R.T. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structure-Activity Relationship of Flavin Analogues That Target the Flavin Mononucleotide Riboswitch.
ACS Chem. Biol., 13, 2018
|
|
3CMH
 
 | | SYNTHETIC LINEAR TRUNCATED ENDOTHELIN-1 AGONIST | | Descriptor: | PROTEIN (ENDOTHELIN-1) | | Authors: | Hewage, C.M, Jiang, L, Parkinson, J.A, Ramage, R, Sadler, I.H. | | Deposit date: | 1998-09-03 | | Release date: | 1999-09-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a novel ETB receptor selective agonist ET1-21 [Cys(Acm)1,15, Aib3,11, Leu7] by nuclear magnetic resonance spectroscopy and molecular modelling.
J.Pept.Res., 53, 1999
|
|
5EY2
 
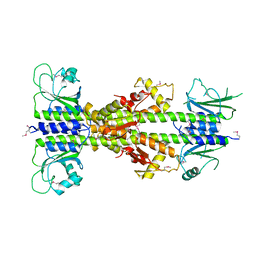 | | Crystal structure of CodY from Bacillus cereus | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY | | Authors: | Han, A, Lee, W.C, Son, J, Kim, S.H, Hwang, K.Y. | | Deposit date: | 2015-11-24 | | Release date: | 2016-09-14 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the pleiotropic transcription regulator CodY provides insight into its GTP-sensing mechanism
Nucleic Acids Res., 44, 2016
|
|
4G4X
 
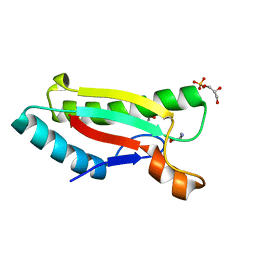 | | Crystal structure of peptidoglycan-associated lipoprotein from Acinetobacter baumannii | | Descriptor: | D-ALANINE, GLYCEROL, Peptidoglycan-associated lipoprotein, ... | | Authors: | Lee, W.C, Song, J.H, Park, J.S, Kim, H.Y. | | Deposit date: | 2012-07-16 | | Release date: | 2013-07-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enantiomer-dependent amino acid binding affinity of OmpA-like domains from Acinetobacter baumannii peptidoglycan-associated lipoprotein and OmpA
To be Published
|
|
4G88
 
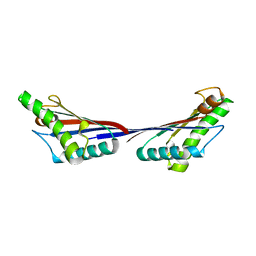 | | Crystal structure of OmpA peptidoglycan-binding domain from Acinetobacter baumannii | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, Outer membrane protein Omp38, S,R MESO-TARTARIC ACID | | Authors: | Lee, W.C, Song, J.H, Park, J.S, Kim, H.Y. | | Deposit date: | 2012-07-22 | | Release date: | 2013-07-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enantiomer-dependent amino acid binding affinity of OmpA-like domains from Acinetobacter baumannii peptidoglycan-associated lipoprotein and OmpA
to be published
|
|
7S0U
 
 | | PRMT5/MEP50 crystal structure with MTA and phthalazinone fragment bound | | Descriptor: | 1,2-ETHANEDIOL, 4-(aminomethyl)phthalazin-1(2H)-one, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ... | | Authors: | Gunn, R.J, Thomas, N.C, Lawson, J.D, Ivetac, A, Smith, C.R, Kulyk, S, Marx, M.A. | | Deposit date: | 2021-08-31 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Fragment-Based Discovery of MRTX1719, a Synthetic Lethal Inhibitor of the PRMT5•MTA Complex for the Treatment of MTAP -Deleted Cancers.
J.Med.Chem., 65, 2022
|
|
7S1Q
 
 | | PRMT5/MEP50 crystal structure with MTA and a phthalazinone inhibitor bound (Compound 9) | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein 50, Protein arginine N-methyltransferase 5, ... | | Authors: | Gunn, R.J, Thomas, N.C, Lawson, J.D, Ivetac, A, Smith, C.R, Kulyk, S, Marx, M.A. | | Deposit date: | 2021-09-02 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Fragment-Based Discovery of MRTX1719, a Synthetic Lethal Inhibitor of the PRMT5•MTA Complex for the Treatment of MTAP -Deleted Cancers.
J.Med.Chem., 65, 2022
|
|
7SER
 
 | | PRMT5/MEP50 with compound 30 bound | | Descriptor: | (2M)-2-{4-[4-(aminomethyl)-1-oxo-1,2-dihydrophthalazin-6-yl]-1-methyl-1H-pyrazol-5-yl}-1-benzothiophene-3-carbonitrile, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein 50, ... | | Authors: | Gunn, R.J, Thomas, N.C, Lawson, J.D, Ivetac, A, Smith, C.R, Kulyk, S, Marx, M.A. | | Deposit date: | 2021-10-01 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Fragment-Based Discovery of MRTX1719, a Synthetic Lethal Inhibitor of the PRMT5•MTA Complex for the Treatment of MTAP -Deleted Cancers.
J.Med.Chem., 65, 2022
|
|
