3PKP
 
 | | Q83S Variant of S. Enterica RmlA with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Glucose-1-phosphate thymidylyltransferase, MAGNESIUM ION | | Authors: | Chang, A, Moretti, R, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Expanding the Nucleotide and Sugar 1-Phosphate Promiscuity of Nucleotidyltransferase RmlA via Directed Evolution.
J.Biol.Chem., 286, 2011
|
|
4K6L
 
 | | Structure of Typhoid Toxin | | Descriptor: | Cytolethal distending toxin subunit B homolog, GLYCEROL, Putative pertussis-like toxin subunit | | Authors: | Gao, X, Song, J, Galan, J. | | Deposit date: | 2013-04-16 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structure and function of the Salmonella Typhi chimaeric A(2)B(5) typhoid toxin.
Nature, 499, 2013
|
|
4L7I
 
 | | Crystal structure of S-Adenosylmethionine synthase from Sulfolobus solfataricus complexed with SAM and PPi | | Descriptor: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-06-13 | | Release date: | 2013-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
2MUS
 
 | | HADDOCK calculated model of LIN5001 bound to the HET-s amyloid | | Descriptor: | 3''',4'-bis(carboxymethyl)-2,2':5',2'':5'',2''':5''',2''''-quinquethiophene-5,5''''-dicarboxylic acid, Heterokaryon incompatibility protein s | | Authors: | Hermann, U.S, Schuetz, A.K, Shirani, H, Saban, D, Nuvolone, M, Huang, D.H, Li, B, Ballmer, B, Aslund, A.K.O, Mason, J.J, Rushing, E, Budka, H, Hammarstrom, P, Bockmann, A, Caflisch, A, Meier, B.H, Nilsson, P.K.R, Hornemann, S, Aguzzi, A. | | Deposit date: | 2014-09-16 | | Release date: | 2017-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-based drug design identifies polythiophenes as antiprion compounds.
Sci Transl Med, 7, 2015
|
|
4PL3
 
 | | Crystal structure of murine IRE1 in complex with MKC9989 inhibitor | | Descriptor: | 7-hydroxy-6-methoxy-3-[2-(2-methoxyethoxy)ethyl]-4,8-dimethyl-2H-chromen-2-one, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sanches, M, Duffy, N, Talukdar, M, Thevakumaran, N, Chiovitti, D, Al-awar, R, Patterson, J.B, Sicheri, F. | | Deposit date: | 2014-05-16 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and mechanism of action of the hydroxy-aryl-aldehyde class of IRE1 endoribonuclease inhibitors.
Nat Commun, 5, 2014
|
|
4IPL
 
 | | The crystal structure of 6-phospho-beta-glucosidase BglA-2 from Streptococcus pneumoniae | | Descriptor: | 6-phospho-beta-glucosidase, GLYCEROL | | Authors: | Yu, W.L, Jiang, Y.L, Andreas, P, Cheng, W, Bai, X.H, Ren, Y.M, Thompsonn, J, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2013-01-10 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structural insights into the substrate specificity of a 6-phospho-&[beta]-glucosidase BglA-2 from Streptococcus pneumoniae TIGR4
J.Biol.Chem., 288, 2013
|
|
4NKD
 
 | | Crystal structure of engineered anti-EE scFv antibody fragment | | Descriptor: | Engineered scFv | | Authors: | Kalyoncu, S, Hyun, J, Pai, J.C, Johnson, J.L, Etzminger, K, Jain, A, Heaner Jr, D, Molares, I.A, Truskett, T.M, Maynard, J.A, Lieberman, R.L. | | Deposit date: | 2013-11-12 | | Release date: | 2014-03-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.303 Å) | | Cite: | Effects of protein engineering and rational mutagenesis on crystal lattice of single chain antibody fragments.
Proteins, 82, 2014
|
|
3CKH
 
 | | Crystal structure of Eph A4 receptor | | Descriptor: | Ephrin type-A receptor 4 | | Authors: | Shi, J.H, Song, J.X. | | Deposit date: | 2008-03-15 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure and NMR Binding Reveal That Two Small Molecule Antagonists Target the High Affinity Ephrin-binding Channel of the EphA4 Receptor.
J.Biol.Chem., 283, 2008
|
|
4PL4
 
 | | Crystal structure of murine IRE1 in complex with OICR464 inhibitor | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, 2-methoxy-6-methyl-4-(4-methyl-3,4-dihydro-2H-1,4-benzoxazin-7-yl)phenol, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sanches, M, Duffy, N, Talukdar, M, Thevakumaran, N, Chiovitti, D, Al-awar, R, Patterson, J.B, Sicheri, F. | | Deposit date: | 2014-05-16 | | Release date: | 2014-09-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and mechanism of action of the hydroxy-aryl-aldehyde class of IRE1 endoribonuclease inhibitors.
Nat Commun, 5, 2014
|
|
3CMH
 
 | | SYNTHETIC LINEAR TRUNCATED ENDOTHELIN-1 AGONIST | | Descriptor: | PROTEIN (ENDOTHELIN-1) | | Authors: | Hewage, C.M, Jiang, L, Parkinson, J.A, Ramage, R, Sadler, I.H. | | Deposit date: | 1998-09-03 | | Release date: | 1999-09-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a novel ETB receptor selective agonist ET1-21 [Cys(Acm)1,15, Aib3,11, Leu7] by nuclear magnetic resonance spectroscopy and molecular modelling.
J.Pept.Res., 53, 1999
|
|
4MQJ
 
 | |
4MQK
 
 | |
4BUP
 
 | | A novel route to product specificity in the Suv4-20 family of histone H4K20 methyltransferases | | Descriptor: | GLYCEROL, HISTONE-LYSINE N-METHYLTRANSFERASE SUV420H1, S-ADENOSYLMETHIONINE, ... | | Authors: | Southall, S.M, Cronin, N.B, Wilson, J.R. | | Deposit date: | 2013-06-21 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.166 Å) | | Cite: | A Novel Route to Product Specificity in the Suv4-20 Family of Histone H4K20 Methyltransferases.
Nucleic Acids Res., 42, 2014
|
|
3QTF
 
 | | Design and SAR of macrocyclic Hsp90 inhibitors with increased metabolic stability and potent cell-proliferation activity | | Descriptor: | (6S)-6,15,15,18-tetramethyl-17-oxo-2,3,4,5,6,7,14,15,16,17-decahydro-1H-8,12-(metheno)[1,4,9]triazacyclotetradecino[9,8-a]indole-9-carboxamide, DIMETHYL SULFOXIDE, Heat shock protein HSP 90-alpha | | Authors: | Zapf, C.W, Bloom, J.D, McBean, J.L, Dushin, R.G, Nittoli, T, Ingalls, C, Sutherland, A.G, Sonye, J.P, Eid, C.N, Golas, J, Liu, H, Boschelli, F, Hu, Y, Vogan, E.M, Levin, J.I. | | Deposit date: | 2011-02-22 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5703 Å) | | Cite: | Design and SAR of macrocyclic Hsp90 inhibitors with increased metabolic stability and potent cell-proliferation activity.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5EEH
 
 | | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with SAH and 2-chloro-4-nitrophenol | | Descriptor: | 2-chloranyl-4-nitro-phenol, Carminomycin 4-O-methyltransferase DnrK, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Wang, F, Singh, S, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-10-22 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Functional AdoMet Isosteres Resistant to Classical AdoMet Degradation Pathways.
Acs Chem.Biol., 11, 2016
|
|
3IKK
 
 | | Crystal structure analysis of msp domain | | Descriptor: | Vesicle-associated membrane protein-associated protein B/C | | Authors: | Shi, J, Lua, S, Song, J. | | Deposit date: | 2009-08-06 | | Release date: | 2010-05-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Elimination of the native structure and solubility of the hVAPB MSP domain by the Pro56Ser mutation that causes amyotrophic lateral sclerosis.
Biochemistry, 49, 2010
|
|
4L2Z
 
 | | Crystal structure of S-Adenosylmethionine synthetase from Sulfolobus solfataricus complexed with SAE and PPi | | Descriptor: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-06-05 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
4NKM
 
 | | Crystal structure of engineered anti-EE scFv antibody fragment | | Descriptor: | Engineered scFv | | Authors: | Kalyoncu, S, Hyun, J, Pai, J.C, Johnson, J.L, Etzminger, K, Jain, A, Heaner Jr, D, Molares, I.A, Truskett, T.M, Maynard, J.A, Lieberman, R.L. | | Deposit date: | 2013-11-12 | | Release date: | 2014-03-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Effects of protein engineering and rational mutagenesis on crystal lattice of single chain antibody fragments.
Proteins, 82, 2014
|
|
5EEG
 
 | | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with tetrazole-SAH | | Descriptor: | (2~{R},3~{R},4~{S},5~{S})-2-(6-aminopurin-9-yl)-5-[[(3~{S})-3-azanyl-3-(1~{H}-1,2,3,4-tetrazol-5-yl)propyl]sulfanylmethyl]oxolane-3,4-diol, Carminomycin 4-O-methyltransferase DnrK | | Authors: | Wang, F, Singh, S, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-10-22 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | Functional AdoMet Isosteres Resistant to Classical AdoMet Degradation Pathways.
Acs Chem.Biol., 11, 2016
|
|
3H78
 
 | | Crystal structure of Pseudomonas aeruginosa PqsD C112A mutant in complex with anthranilic acid | | Descriptor: | 2-AMINOBENZOIC ACID, PQS biosynthetic enzyme | | Authors: | Bera, A.K, Atanasova, V, Parsons, J.F. | | Deposit date: | 2009-04-24 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of PqsD, a Pseudomonas quinolone signal biosynthetic enzyme, in complex with anthranilate.
Biochemistry, 48, 2009
|
|
3H76
 
 | | Crystal structure of PqsD, a key enzyme in Pseudomonas aeruginosa quinolone signal biosynthesis pathway | | Descriptor: | PQS biosynthetic enzyme | | Authors: | Bera, A.K, Atanasova, V, Parsons, J.F. | | Deposit date: | 2009-04-24 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of PqsD, a Pseudomonas quinolone signal biosynthetic enzyme, in complex with anthranilate.
Biochemistry, 48, 2009
|
|
3JD0
 
 | | Glutamate dehydrogenase in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3DDX
 
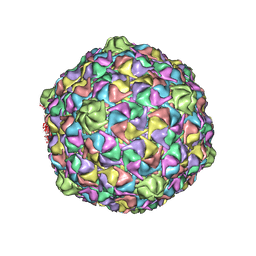 | | HK97 bacteriophage capsid Expansion Intermediate-II model | | Descriptor: | Major capsid protein | | Authors: | Lee, K.K, Gan, L, Conway, J.F, Hendrix, R.W, Steven, A.C, Johnson, J.E. | | Deposit date: | 2008-06-06 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY | | Cite: | Virus capsid expansion driven by the capture of mobile surface loops.
Structure, 16, 2008
|
|
3JD4
 
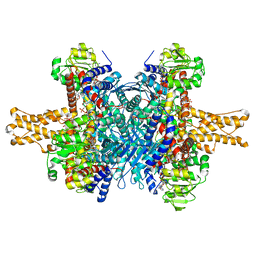 | | Glutamate dehydrogenase in complex with NADH and GTP, closed conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, ... | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3DVF
 
 | |
