5UG1
 
 | | Structure of Streptococcus pneumoniae peptidoglycan O-acetyltransferase A (OatA) C-terminal catalytic domain with methylsulfonyl adduct | | Descriptor: | Acyltransferase, SODIUM ION, methanesulfonic acid | | Authors: | Sychantha, D, Jones, C, Little, D.J, Moynihan, P.J, Robinson, H, Galley, N.F, Roper, D.I, Dowson, C.G, Howell, P.L, Clarke, A.J. | | Deposit date: | 2017-01-06 | | Release date: | 2017-10-25 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | In vitro characterization of the antivirulence target of Gram-positive pathogens, peptidoglycan O-acetyltransferase A (OatA).
PLoS Pathog., 13, 2017
|
|
5UFY
 
 | | Structure of Streptococcus pneumoniae peptidoglycan O-acetyltransferase A (OatA) C-terminal catalytic domain | | Descriptor: | Acyltransferase, SODIUM ION | | Authors: | Sychantha, D, Jones, C, Little, D.J, Moynihan, P.J, Robinson, H, Galley, N.F, Roper, D.I, Dowson, C.G, Howell, P.L, Clarke, A.J. | | Deposit date: | 2017-01-06 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | In vitro characterization of the antivirulence target of Gram-positive pathogens, peptidoglycan O-acetyltransferase A (OatA).
PLoS Pathog., 13, 2017
|
|
3Q5D
 
 | |
3PO0
 
 | | Crystal structure of SAMP1 from Haloferax volcanii | | Descriptor: | ACETATE ION, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Jeong, Y.J, Jeong, B.-C, Song, H.K. | | Deposit date: | 2010-11-21 | | Release date: | 2011-03-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of ubiquitin-like small archaeal modifier protein 1 (SAMP1) from Haloferax volcanii.
Biochem.Biophys.Res.Commun., 405, 2011
|
|
5TCB
 
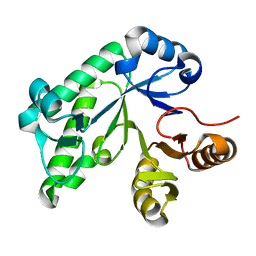 | |
1TEG
 
 | | Crystal structure of the spinach plastocyanin mutants G8D/K30C/T69C and K30C/T69C- a study of the effect on crystal packing and thermostability from the introduction of a novel disulfide bond | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Plastocyanin, ... | | Authors: | Okvist, M, Jacobson, F, Jansson, H, Hansson, O, Sjolin, L. | | Deposit date: | 2004-05-25 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Novel Disulfide Bonds Effect the Thermostability of Plastocyanin. Crystal structures of the triple plastocyanin
mutant G8D/K30C/T69C and the double plastocyanin mutant K30C/T69C from spinach at 1.90 A and 1.96 A resolution, respectively.
To be Published
|
|
4ATW
 
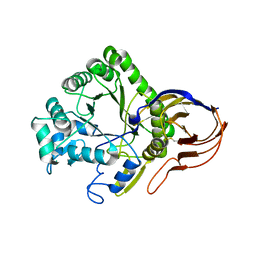 | | The crystal structure of Arabinofuranosidase | | Descriptor: | ALPHA-L-ARABINOFURANOSIDASE DOMAIN PROTEIN | | Authors: | Dumbrepatil, A, Song, H.-N, Jung, T.-Y, Kim, T.-J, Woo, E.-J. | | Deposit date: | 2012-05-10 | | Release date: | 2012-05-23 | | Last modified: | 2013-01-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Analysis of Alpha-L-Arabinofuranosidase from Thermotoga Maritima Reveals Characteristics for Thermostability and Substrate Specificity.
J.Microbiol.Biotech., 22, 2012
|
|
4DN0
 
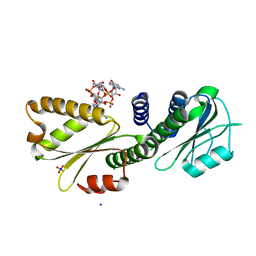 | | PelD 156-455 from Pseudomonas aeruginosa PA14 in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Putative uncharacterized protein pelD, SODIUM ION, ... | | Authors: | Whitney, J.C, Colvin, K.M, Marmont, L.S, Robinson, H, Parsek, M.R, Howell, P.L. | | Deposit date: | 2012-02-08 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Cytoplasmic Region of PelD, a Degenerate Diguanylate Cyclase Receptor That Regulates Exopolysaccharide Production in Pseudomonas aeruginosa.
J.Biol.Chem., 287, 2012
|
|
4HU8
 
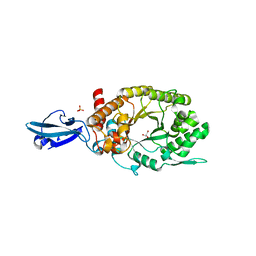 | | Crystal Structure of a Bacterial Ig-like Domain Containing GH10 Xylanase from Termite Gut | | Descriptor: | GH10 Xylanase, GLYCEROL, SULFATE ION | | Authors: | Han, Q, Liu, N, Robinson, H, Cao, L, Qian, C, Wang, Q, Xie, L, Ding, H, Wang, Q, Huang, Y, Li, J, Zhou, Z. | | Deposit date: | 2012-11-02 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical characterization and crystal structure of a GH10 xylanase from termite gut bacteria reveal a novel structural feature and significance of its bacterial Ig-like domain.
Biotechnol.Bioeng., 110, 2013
|
|
1TEF
 
 | | Crystal structure of the spinach plastocyanin mutants G8D/K30C/T69C and K30C/T69C- a study of the effect on crystal packing and thermostability from the introduction of a novel disulfide bond | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Plastocyanin, ... | | Authors: | Okvist, M, Jacobson, F, Jansson, H, Hansson, O, Sjolin, L. | | Deposit date: | 2004-05-25 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel Disulfide Bonds Effect the Thermostability of Plastocyanin. Crystal structures of the triple plastocyanin mutant G8D/K30C/T69C and the double plastocyanin mutant K30C/T69C from spinach at 1.90 and 1.96 resolution, respectively.
To be Published
|
|
1QHF
 
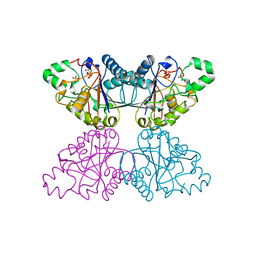 | | YEAST PHOSPHOGLYCERATE MUTASE-3PG COMPLEX STRUCTURE TO 1.7 A | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, PROTEIN (PHOSPHOGLYCERATE MUTASE), SULFATE ION | | Authors: | Crowhurst, G, Littlechild, J, Watson, H.C. | | Deposit date: | 1999-05-13 | | Release date: | 1999-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a phosphoglycerate mutase:3-phosphoglyceric acid complex at 1.7 A.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2GK6
 
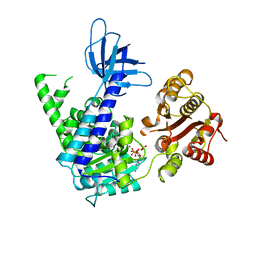 | | Structural and Functional insights into the human Upf1 helicase core | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Cheng, Z, Muhlrad, D, Parker, R, Song, H. | | Deposit date: | 2006-03-31 | | Release date: | 2007-01-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insights into the human Upf1 helicase core
Embo J., 26, 2007
|
|
1RGO
 
 | | Structural Basis for Recognition of the mRNA Class II AU-Rich Element by the Tandem Zinc Finger Domain of TIS11d | | Descriptor: | Butyrate response factor 2, RNA (5'-R(*UP*UP*AP*UP*UP*UP*AP*UP*U)-3'), ZINC ION | | Authors: | Hudson, B.P, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2003-11-12 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d.
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
4E14
 
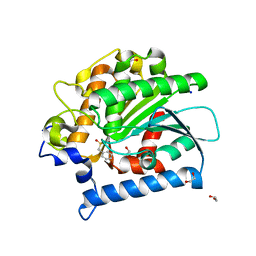 | | Crystal structure of kynurenine formamidase conjugated with phenylmethylsulfonyl fluoride | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, kynurenine formamidase | | Authors: | Han, Q, Robinson, H, Li, J. | | Deposit date: | 2012-03-05 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Biochemical identification and crystal structure of kynurenine formamidase from Drosophila melanogaster.
Biochem.J., 446, 2012
|
|
1UCS
 
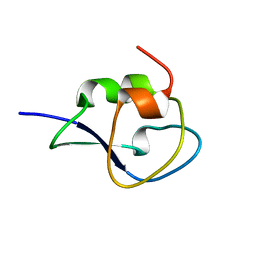 | | Type III Antifreeze Protein RD1 from an Antarctic Eel Pout | | Descriptor: | Antifreeze peptide RD1 | | Authors: | Ko, T.-P, Robinson, H, Gao, Y.-G, Cheng, C.-H.C, DeVries, A.L, Wang, A.H.-J. | | Deposit date: | 2003-04-21 | | Release date: | 2003-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.62 Å) | | Cite: | The refined crystal structure of an eel pout type III antifreeze protein RD1 at 0.62-A resolution reveals structural microheterogeneity of protein and solvation.
Biophys.J., 84, 2003
|
|
2OUN
 
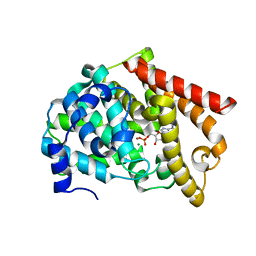 | | crystal structure of PDE10A2 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H, Ke, H.M. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2EWG
 
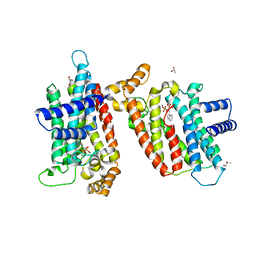 | | T. brucei Farnesyl Diphosphate Synthase Complexed with Minodronate | | Descriptor: | (1-HYDROXY-2-IMIDAZO[1,2-A]PYRIDIN-3-YLETHANE-1,1-DIYL)BIS(PHOSPHONIC ACID), MAGNESIUM ION, S-1,2-PROPANEDIOL, ... | | Authors: | Cao, R, Mao, J, Gao, Y, Robinson, H, Odeh, S, Goddard, A, Oldfield, E. | | Deposit date: | 2005-11-03 | | Release date: | 2006-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Solid-state NMR, crystallographic, and computational investigation of bisphosphonates and farnesyl diphosphate synthase-bisphosphonate complexes.
J.Am.Chem.Soc., 128, 2006
|
|
4FWY
 
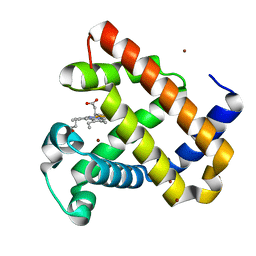 | | F33Y CuB myoglobin (F33Y L29H F43H sperm whale myoglobin) with copper bound | | Descriptor: | COPPER (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gao, Y.-G, Robinson, H, Petrik, I.D, Miner, K.D, Lu, Y. | | Deposit date: | 2012-07-02 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Designed Functional Metalloenzyme that Reduces O(2) to H(2) O with Over One Thousand Turnovers.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
1R7S
 
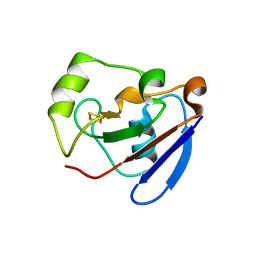 | | PUTIDAREDOXIN (Fe2S2 ferredoxin), C73G mutant | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Putidaredoxin | | Authors: | Smith, N, Mayhew, M, Kelly, H, Robinson, H, Heroux, A, Holden, M.J, Gallagher, D.T. | | Deposit date: | 2003-10-22 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of C73G putidaredoxin from Pseudomonas putida.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1SD4
 
 | | Crystal Structure of a SeMet derivative of BlaI at 2.0 A | | Descriptor: | PENICILLINASE REPRESSOR, SULFATE ION | | Authors: | Safo, M.K, Zhao, Q, Musayev, F.N, Robinson, H, Scarsdale, N, Archer, G.L. | | Deposit date: | 2004-02-13 | | Release date: | 2004-08-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the BlaI repressor from Staphylococcus aureus and its complex with DNA: insights into transcriptional regulation of the bla and mec operons
J.Bacteriol., 187, 2005
|
|
1QZY
 
 | | Human Methionine Aminopeptidase in complex with bengamide inhibitor LAF153 and cobalt | | Descriptor: | (E)-(2R,3R,4S,5R)-3,4,5-TRIHYDROXY-2-METHOXY-8,8-DIMETHYL-NON-6-ENOIC ACID ((3S,6R)-6-HYDROXY-2-OXO-AZEPAN-3-YL)-AMIDE, COBALT (II) ION, Methionine aminopeptidase 2, ... | | Authors: | Eck, M.J, Song, H.K, Morollo, A. | | Deposit date: | 2003-09-18 | | Release date: | 2003-11-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Proteomics-based target identification: bengamides as a new class of methionine aminopeptidase inhibitors.
J.Biol.Chem., 278, 2003
|
|
1C8C
 
 | | CRYSTAL STRUCTURES OF THE CHROMOSOMAL PROTEINS SSO7D/SAC7D BOUND TO DNA CONTAINING T-G MISMATCHED BASE PAIRS | | Descriptor: | 5'-D(*GP*TP*GP*AP*TP*CP*GP*C)-3', DNA-BINDING PROTEIN 7A | | Authors: | Su, S, Gao, Y.-G, Robinson, H, Liaw, Y.-C, Edmondson, S.P, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 2000-05-04 | | Release date: | 2001-05-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structures of the chromosomal proteins Sso7d/Sac7d bound to DNA containing T-G mismatched base-pairs.
J.Mol.Biol., 303, 2000
|
|
5U6H
 
 | | Solution structure of the zinc fingers 1 and 2 of MBNL1 | | Descriptor: | Muscleblind-like protein 1, ZINC ION | | Authors: | Phukan, P.D, Park, S, Martinez-Yamout, M.M, Zeeb, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-12-08 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Interaction of the Tandem Zinc Finger Domains of Human Muscleblind with Cognate RNA from Human Cardiac Troponin T.
Biochemistry, 56, 2017
|
|
4HS4
 
 | | Crystal structure of a putative chromate reductase from Gluconacetobacter hansenii, Gh-ChrR, containing a Y129N substitution. | | Descriptor: | Chromate reductase, FLAVIN MONONUCLEOTIDE | | Authors: | Zhang, Y, Robinson, H, Buchko, G.W. | | Deposit date: | 2012-10-29 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic insights of chromate and uranyl reduction by the NADPH-dependent FMN reductase, ChrR, from Gluconacetobacter hansenii
To be Published
|
|
4H6P
 
 | | Crystal structure of a putative chromate reductase from Gluconacetobacter hansenii, Gh-ChrR, containing a R101A substitution. | | Descriptor: | Chromate reductase, FLAVIN MONONUCLEOTIDE | | Authors: | Zhang, Y, Robinson, H, Buchko, G.W. | | Deposit date: | 2012-09-19 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.556 Å) | | Cite: | Mechanistic insights of chromate and uranyl reduction by the NADPH-dependent FMN reductase, ChrR, from Gluconacetobacter hansenii
To be Published
|
|
