3HTS
 
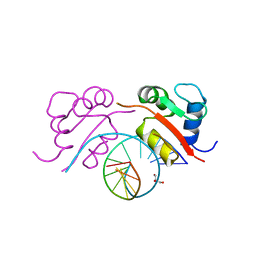 | | HEAT SHOCK TRANSCRIPTION FACTOR/DNA COMPLEX | | Descriptor: | 5'-D(*GP*GP*TP*TP*CP*TP*AP*GP*AP*AP*CP*C)-3', GLYCEROL, HEAT SHOCK TRANSCRIPTION FACTOR | | Authors: | Littlefield, O, Nelson, H.C.M. | | Deposit date: | 1998-11-16 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A new use for the 'wing' of the 'winged' helix-turn-helix motif in the HSF-DNA cocrystal.
Nat.Struct.Biol., 6, 1999
|
|
3HRS
 
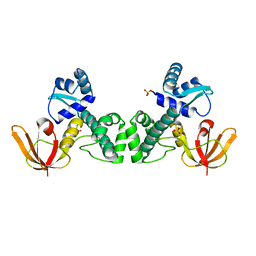 | | Crystal Structure of the Manganese-activated Repressor ScaR: apo form | | Descriptor: | Metalloregulator ScaR, SULFATE ION | | Authors: | Stoll, K.E, Draper, W.E, Kliegman, J.I, Golynskiy, M.V, Brew-Appiah, R.A.T, Brown, H.K, Breyer, W.A, Jakubovics, N.S, Jenkinson, H.F, Brennan, R.B, Cohen, S.M, Glasfeld, A. | | Deposit date: | 2009-06-09 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Characterization and structure of the manganese-responsive transcriptional regulator ScaR.
Biochemistry, 48, 2009
|
|
3MN0
 
 | | Introducing a 2-His-1-Glu Non-Heme Iron Center into Myoglobin confers Nitric Oxide Reductase activity: Cu(II)-CN-FeBMb(-His) form | | Descriptor: | COPPER (II) ION, CYANIDE ION, Myoglobin, ... | | Authors: | Lin, Y.-W, Yeung, N, Gao, Y.-G, Miner, K.D, Lei, L, Robinson, H, Lu, Y. | | Deposit date: | 2010-04-20 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Introducing a 2-his-1-glu nonheme iron center into myoglobin confers nitric oxide reductase activity.
J.Am.Chem.Soc., 132, 2010
|
|
3HRU
 
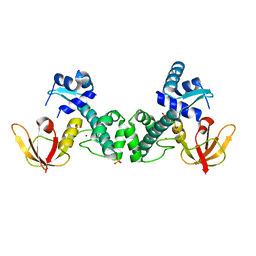 | | Crystal Structure of ScaR with bound Zn2+ | | Descriptor: | Metalloregulator ScaR, SULFATE ION, ZINC ION | | Authors: | Stoll, K.E, Draper, W.E, Kliegman, J.I, Golynskiy, M.V, Brew-Appiah, R.A.T, Brown, H.K, Breyer, W.A, Jakubovics, N.S, Jenkinson, H.F, Brennan, R.B, Cohen, S.M, Glasfeld, A. | | Deposit date: | 2009-06-09 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Characterization and structure of the manganese-responsive transcriptional regulator ScaR.
Biochemistry, 48, 2009
|
|
4KOL
 
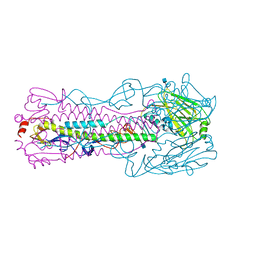 | | The structure of hemagglutinin from avian-origin H7N9 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, Hemagglutinin HA2 | | Authors: | Shi, Y, Zhang, W, Wang, F, Qi, J, Song, H, Wu, Y, Gao, F, Zhang, Y, Fan, Z, Gong, W, Wang, D, Shu, Y, Wang, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-05-12 | | Release date: | 2013-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses.
Science, 342, 2013
|
|
4LSD
 
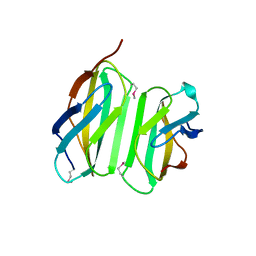 | | Myokine structure | | Descriptor: | Fibronectin type III domain-containing protein 5 | | Authors: | Schumacher, M.A, Ohashi, T, Shah, R.S, Chinnam, N, Erickson, H. | | Deposit date: | 2013-07-22 | | Release date: | 2013-10-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The structure of irisin reveals a novel intersubunit beta-sheet fibronectin type III (FNIII) dimer: implications for receptor activation.
J.Biol.Chem., 288, 2013
|
|
3MCA
 
 | |
3NIH
 
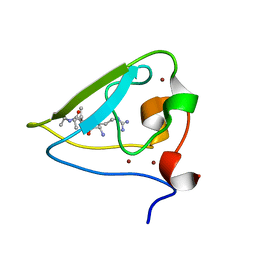 | | The structure of UBR box (RIAAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide RIAAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3CS1
 
 | | Flagellar Calcium-binding Protein (FCaBP) from T. cruzi | | Descriptor: | Flagellar calcium-binding protein | | Authors: | Ames, J.B, Ladner, J.E, Wingard, J.N, Robinson, H, Fisher, A. | | Deposit date: | 2008-04-08 | | Release date: | 2008-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into Membrane Targeting by the Flagellar Calcium-binding Protein (FCaBP), a Myristoylated and Palmitoylated Calcium Sensor in Trypanosoma cruzi.
J.Biol.Chem., 283, 2008
|
|
8POF
 
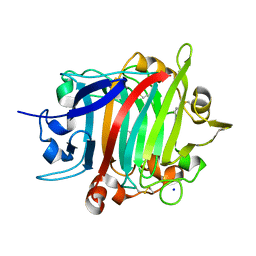 | | The crystal structure of RsSymEG1 reveals a unique form of smaller GH7 endoglucanases alongside GH7 cellobiohydrolases in protist symbionts of termites | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Putative glycosyl hydrolase family7, SODIUM ION | | Authors: | Haataja, T, Sandgren, M, Hansson, H, Stahlberg, J. | | Deposit date: | 2023-07-04 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of RsSymEG1 reveals a unique form of smaller GH7 endoglucanases alongside GH7 cellobiohydrolases in protist symbionts of termites.
Febs J., 291, 2024
|
|
1UCS
 
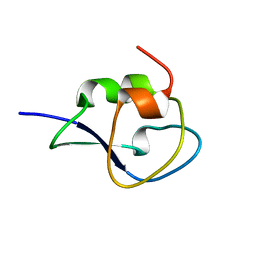 | | Type III Antifreeze Protein RD1 from an Antarctic Eel Pout | | Descriptor: | Antifreeze peptide RD1 | | Authors: | Ko, T.-P, Robinson, H, Gao, Y.-G, Cheng, C.-H.C, DeVries, A.L, Wang, A.H.-J. | | Deposit date: | 2003-04-21 | | Release date: | 2003-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.62 Å) | | Cite: | The refined crystal structure of an eel pout type III antifreeze protein RD1 at 0.62-A resolution reveals structural microheterogeneity of protein and solvation.
Biophys.J., 84, 2003
|
|
1F77
 
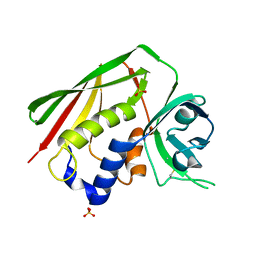 | | STAPHYLOCOCCAL ENTEROTOXIN H DETERMINED TO 2.4 A RESOLUTION | | Descriptor: | ENTEROTOXIN H, SULFATE ION | | Authors: | Hakansson, M, Petersson, K, Nilsson, H, Forsberg, G, Bjork, P. | | Deposit date: | 2000-06-26 | | Release date: | 2000-07-19 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of staphylococcal enterotoxin H: implications for binding properties to MHC class II and TcR molecules.
J.Mol.Biol., 302, 2000
|
|
5HP0
 
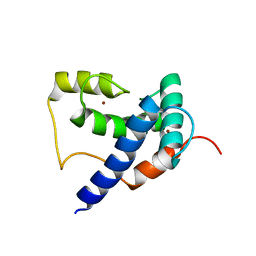 | | Solution Structure of TAZ2-p53AD2 | | Descriptor: | CREB-binding protein,Cellular tumor antigen p53 fusion protein, ZINC ION | | Authors: | Krois, A.S, Ferreon, J.C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3EZQ
 
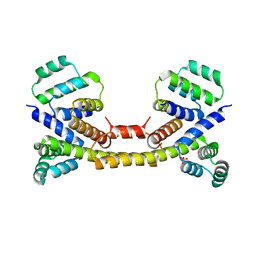 | | Crystal Structure of the Fas/FADD Death Domain Complex | | Descriptor: | Protein FADD, SODIUM ION, SULFATE ION, ... | | Authors: | Schwarzenbacher, R, Robinson, H, Stec, B, Riedl, S.J. | | Deposit date: | 2008-10-23 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The Fas-FADD death domain complex structure unravels signalling by receptor clustering
Nature, 457, 2009
|
|
5HPD
 
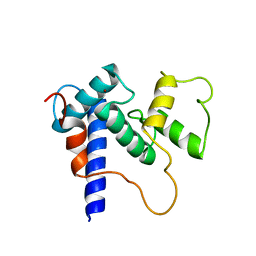 | | Solution Structure of TAZ2-p53TAD | | Descriptor: | CREB-binding protein,Cellular tumor antigen p53 fusion protein, ZINC ION | | Authors: | Krois, A.S, Ferreon, J.C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-01-20 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5HOU
 
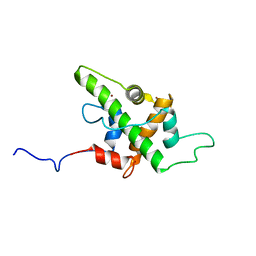 | | Solution Structure of p53TAD-TAZ1 | | Descriptor: | Cellular tumor antigen p53,CREB-binding protein fusion protein, ZINC ION | | Authors: | Krois, A.S, Ferreon, J.C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EOY
 
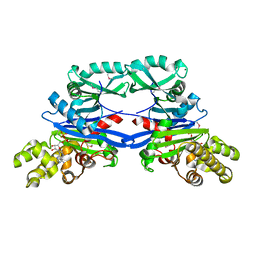 | | Pseudomonas aeruginosa SeMet-PilM bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Type 4 fimbrial biogenesis protein PilM | | Authors: | McCallum, M, Tammam, S, Robinson, H, Shah, M, Calmettes, C, Moraes, T, Burrows, L.L, Howell, P.L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-04-27 | | Last modified: | 2016-06-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | PilN Binding Modulates the Structure and Binding Partners of the Pseudomonas aeruginosa Type IVa Pilus Protein PilM.
J.Biol.Chem., 291, 2016
|
|
5EQ6
 
 | | Pseudomonas aeruginosa PilM bound to AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Type 4 fimbrial biogenesis protein PilM | | Authors: | McCallum, M, Tammam, S, Robinson, H, Shah, M, Calmettes, C, Moraes, T, Burrows, L.L, Howell, P.L. | | Deposit date: | 2015-11-12 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | PilN Binding Modulates the Structure and Binding Partners of the Pseudomonas aeruginosa Type IVa Pilus Protein PilM.
J.Biol.Chem., 291, 2016
|
|
5EOX
 
 | | Pseudomonas aeruginosa PilM bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Type 4 fimbrial biogenesis protein PilM | | Authors: | McCallum, M, Tammam, S, Robinson, H, Shah, M, Calmettes, C, Moraes, T, Burrows, L.L, Howell, P.L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PilN Binding Modulates the Structure and Binding Partners of the Pseudomonas aeruginosa Type IVa Pilus Protein PilM.
J.Biol.Chem., 291, 2016
|
|
5EOU
 
 | | Pseudomonas aeruginosa PilM:PilN1-12 bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | McCallum, M, Tammam, S, Robinson, H, Shah, M, Calmettes, C, Moraes, T, Burrows, L, Howell, L.P. | | Deposit date: | 2015-11-10 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PilN Binding Modulates the Structure and Binding Partners of the Pseudomonas aeruginosa Type IVa Pilus Protein PilM.
J.Biol.Chem., 291, 2016
|
|
5EUH
 
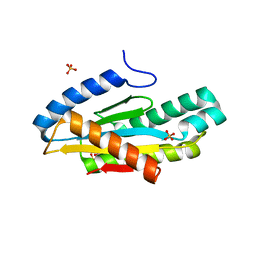 | | Crystal structure of the c-di-GMP-bound GGDEF domain of P. fluorescens GcbC | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Putative GGDEF domain membrane protein, SULFATE ION | | Authors: | Giglio, K.M, Cooley, R.B, Sondermann, H. | | Deposit date: | 2015-11-18 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Contribution of Physical Interactions to Signaling Specificity between a Diguanylate Cyclase and Its Effector.
Mbio, 6, 2015
|
|
4OJX
 
 | | crystal structure of yeast phosphodiesterase-1 in complex with GMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3',5'-cyclic-nucleotide phosphodiesterase 1, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Tian, Y, Cui, W, Huang, M, Robinson, H, Wan, Y, Wang, Y, Ke, H. | | Deposit date: | 2014-01-21 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Dual specificity and novel structural folding of yeast phosphodiesterase-1 for hydrolysis of second messengers cyclic adenosine and guanosine 3',5'-monophosphate.
Biochemistry, 53, 2014
|
|
5EXP
 
 | | AAA+ domain of FleQ from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Transcriptional regulator FleQ | | Authors: | Navarro, M.V.A.S, Sondermann, H, Matsuyama, B.Y. | | Deposit date: | 2015-11-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanistic insights into c-di-GMP-dependent control of the biofilm regulator FleQ from Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4B68
 
 | | A. fumigatus ornithine hydroxylase (SidA), re-oxidised state bound to NADP and Arg | | Descriptor: | ARGININE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Franceschini, S, Fedkenheuer, M, Vogelaar, N.J, Robinson, H.H, Sobrado, P, Mattevi, A. | | Deposit date: | 2012-08-09 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural Insight Into the Mechanism of Oxygen Activation and Substrate Selectivity of Flavin-Dependent N-Hydroxylating Monooxygenases.
Biochemistry, 51, 2012
|
|
4B64
 
 | | A. fumigatus ornithine hydroxylase (SidA) bound to NADP and Lysine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, L-ORNITHINE N5 MONOOXYGENASE, ... | | Authors: | Franceschini, S, Fedkenheuer, M, Vogelaar, N.J, Robinson, H.H, Sobrado, P, Mattevi, A. | | Deposit date: | 2012-08-09 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Insight Into the Mechanism of Oxygen Activation and Substrate Selectivity of Flavin-Dependent N-Hydroxylating Monooxygenases.
Biochemistry, 51, 2012
|
|
