1BD2
 
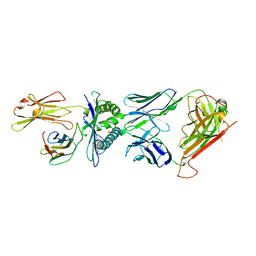 | | COMPLEX BETWEEN HUMAN T-CELL RECEPTOR B7, VIRAL PEPTIDE (TAX) AND MHC CLASS I MOLECULE HLA-A 0201 | | Descriptor: | BETA-2 MICROGLOBULIN, HLA-A 0201, T CELL RECEPTOR ALPHA, ... | | Authors: | Ding, Y.-H, Smith, K.J, Garboczi, D.N, Utz, U, Biddison, W.E, Wiley, D.C. | | Deposit date: | 1998-05-12 | | Release date: | 1998-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two human T cell receptors bind in a similar diagonal mode to the HLA-A2/Tax peptide complex using different TCR amino acids.
Immunity, 8, 1998
|
|
2BM2
 
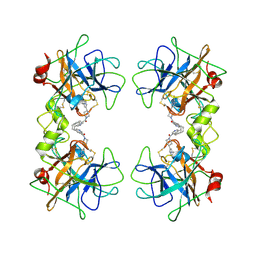 | | human beta-II tryptase in complex with 4-(3-Aminomethyl-phenyl)- piperidin-1-yl-(5-phenethyl- pyridin-3-yl)-methanone | | Descriptor: | 1-[3-(1-{[5-(2-PHENYLETHYL)PYRIDIN-3-YL]CARBONYL}PIPERIDIN-4-YL)PHENYL]METHANAMINE, HUMAN BETA2 TRYPTASE | | Authors: | Maignan, S, Guilloteau, J.-P, Dupuy, A, Levell, J, Astles, P, Eastwood, P, Cairns, J, Houille, O, Aldous, S, Merriman, G, Whiteley, B, Pribish, J, Czekaj, M, Liang, G, Davidson, J, Harrison, T, Morley, A, Watson, S, Fenton, G, Mccarthy, C, Romano, J, Mathew, R, Engers, D, Gardyan, M, Sides, K, Kwong, J, Tsay, J, Rebello, S, Shen, L, Wang, J, Luo, Y, Giardino, O, Lim, H.-K, Smith, K, Pauls, H. | | Deposit date: | 2005-03-09 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Based Design of 4-(3-Aminomethylphenyl) Piperidinyl-1-Amides: Novel, Potent, Selective, and Orally Bioavailable Inhibitors of Bii Tryptase
Bioorg.Med.Chem., 13, 2005
|
|
1G5C
 
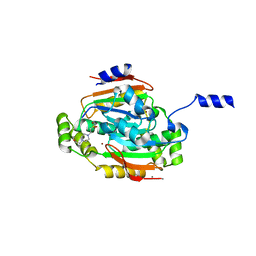 | | CRYSTAL STRUCTURE OF THE 'CAB' TYPE BETA CLASS CARBONIC ANHYDRASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-CARBONIC ANHYDRASE, CALCIUM ION, ... | | Authors: | Strop, P, Smith, K.S, Iverson, T.M, Ferry, J.G, Rees, D.C. | | Deposit date: | 2000-10-31 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the "cab"-type beta class carbonic anhydrase from the archaeon Methanobacterium thermoautotrophicum.
J.Biol.Chem., 276, 2001
|
|
1E3T
 
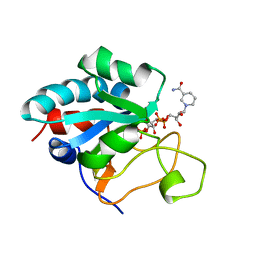 | | Solution Structure of the NADP(H) binding Component (dIII) of Proton-Translocating Transhydrogenase from Rhodospirillum rubrum | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICOTINAMIDE NUCLEOTIDE TRANSHYDROGENASE (SUBUNIT BETA) | | Authors: | Jeeves, M, Smith, K.J, Quirk, P.G, Cotton, N.P.J, Jackson, J.B. | | Deposit date: | 2000-06-22 | | Release date: | 2000-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Nadp(H)-Binding Component (Diii) of Proton-Translocating Transhydrogenase from Rhodospirillum Rubrum
Biochim.Biophys.Acta, 1459, 2000
|
|
3J05
 
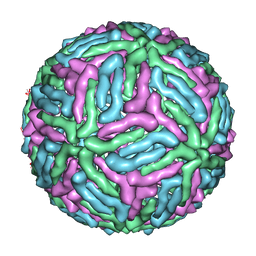 | | Three-dimensional structure of Dengue virus serotype 1 complexed with HMAb 14c10 Fab | | Descriptor: | envelope protein | | Authors: | Teoh, E.P, Kukkaro, P, Teo, E.W, Lim, A, Tan, T.T, Shi, P.Y, Yip, A, Schul, W, Leo, Y.S, Chan, S.H, Smith, K.G.C, Ooi, E.E, Kemeny, D.M, Ng, G, Ng, M.L, Alonso, S, Fisher, D, Hanson, B, Lok, S.M, MacAry, P.A. | | Deposit date: | 2011-04-01 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | The structural basis for serotype-specific neutralization of dengue virus by a human antibody.
Sci Transl Med, 4, 2012
|
|
1E9K
 
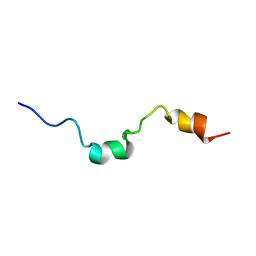 | | The structure of the RACK1 interaction sites located within the unique N-terminal region of the cAMP-specific phosphodiesterase, PDE4D5. | | Descriptor: | cAMP-specific 3',5'-cyclic phosphodiesterase 4D | | Authors: | Bolger, G.B, Smith, K.J, McCahill, A, Hyde, E.I, Steele, M.R, Houslay, M.D. | | Deposit date: | 2000-10-20 | | Release date: | 2001-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H NMR structural and functional characterisation of a cAMP-specific phosphodiesterase-4D5 (PDE4D5) N-terminal region peptide that disrupts PDE4D5 interaction with the signalling scaffold proteins, beta-arrestin and RACK1.
Cell. Signal., 19, 2007
|
|
4CAU
 
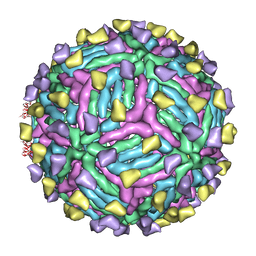 | | THREE-DIMENSIONAL STRUCTURE OF DENGUE VIRUS SEROTYPE 1 COMPLEXED WITH 2 HMAB 14C10 FAB | | Descriptor: | ENVELOPE PROTEIN E, FAB 14C10 | | Authors: | Teoh, E.P, Kukkaro, P, Teo, E.W, Lim, A.P, Tan, T.T, Yip, A, Schul, W, Aung, M, Kostyuchenko, V.A, Leo, Y.S, Chan, S.H, Smith, K.G, Chan, A.H, Zou, G, Ooi, E.E, Kemeny, D.M, Tan, G.K, Ng, J.K, Ng, M.L, Alonso, S, Fisher, D, Shi, P.Y, Hanson, B.J, Lok, S.M, Macary, P.A. | | Deposit date: | 2013-10-09 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | The Structural Basis for Serotype-Specific Neutralization of Dengue Virus by a Human Antibody.
Sci.Trans.Med, 4, 2012
|
|
3G8T
 
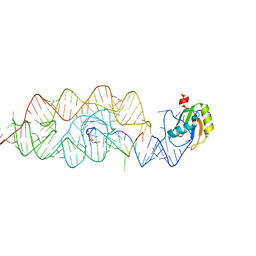 | | Crystal structure of the G33A mutant Bacillus anthracis glmS ribozyme bound to GlcN6P | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, RNA (5'-R(*AP*(A2M)P*GP*CP*GP*CP*CP*AP*GP*AP*AP*CP*U)-3'), ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-12 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
3G9C
 
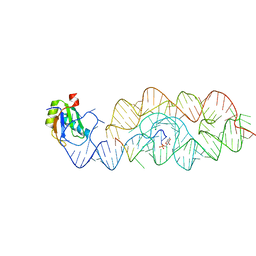 | | Crystal structure of the product Bacillus anthracis glmS ribozyme | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, GLMS RIBOZYME, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-13 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
3G96
 
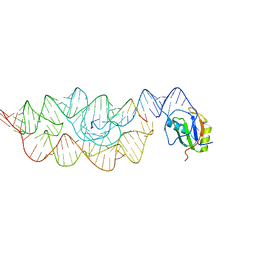 | | Crystal structure of the Bacillus anthracis glmS ribozyme bound to MaN6P | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-mannopyranose, GLMS RIBOZYME, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-12 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
2PR9
 
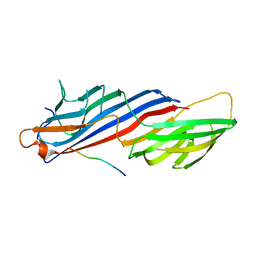 | | Mu2 adaptin subunit (AP50) of AP2 adaptor (second domain), complexed with GABAA receptor-gamma2 subunit-derived internalization peptide DEEYGYECL | | Descriptor: | AP-2 complex subunit mu-1, GABA(A) receptor subunit gamma-2 peptide | | Authors: | Vahedi-Faridi, A, Haucke, V, Kittler, J.T, Kukhtina, V, Moss, S.J, Saenger, W, Chen, G.-J, Tretter, V, Smith, K, Yan, Z, McAinsh, K, Arancibia-Carcamo, L. | | Deposit date: | 2007-05-04 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Regulation of synaptic inhibition by phospho-dependent binding of the AP2 complex to a YECL motif in the GABAA receptor gamma2 subunit.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7KRM
 
 | |
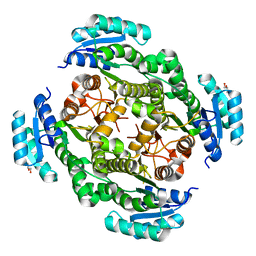
7KRK
 
 | |
3L3C
 
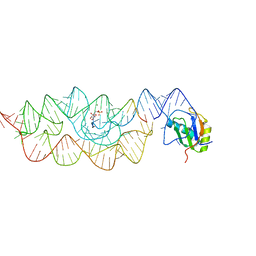 | | Crystal structure of the Bacillus anthracis glmS ribozyme bound to Glc6P | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, GLMS RIBOZYME, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-12-16 | | Release date: | 2009-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
4BIB
 
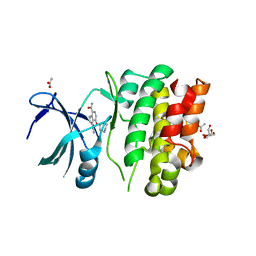 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | 3-cyano-4-(piperidin-4-yloxy)-1H-indole-7-carboxamide, ACETATE ION, GLYCEROL, ... | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
3MXH
 
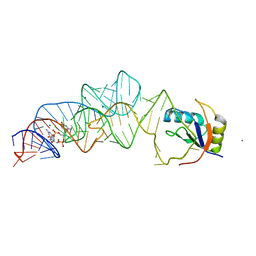 | | Native structure of a c-di-GMP riboswitch from V. cholerae | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, U1 small nuclear ribonucleoprotein A, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-07 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
3MUT
 
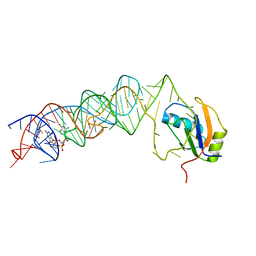 | | Crystal Structure of the G20A/C92U mutant c-di-GMP riboswith bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), G20A/C92U mutant c-di-GMP riboswitch, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
3MUR
 
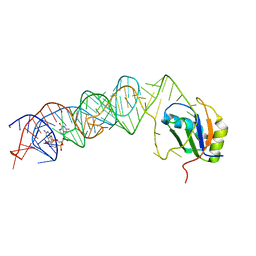 | | Crystal Structure of the C92U mutant c-di-GMP riboswith bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), C92U mutant c-di-GMP riboswitch, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
3MUM
 
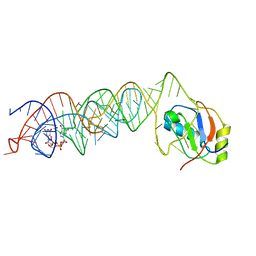 | | Crystal Structure of the G20A mutant c-di-GMP riboswith bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), G20A mutant c-di-GMP Riboswitch, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
3MUV
 
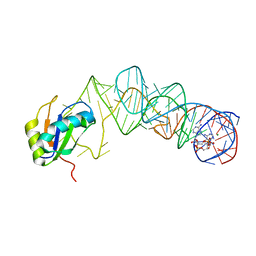 | | Crystal Structure of the G20A/C92U mutant c-di-GMP riboswith bound to c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, G20A/C92U mutant c-di-GMP riboswitch, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
3G8S
 
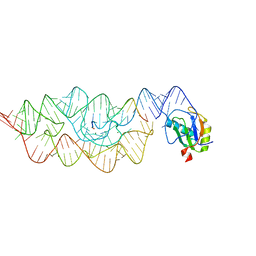 | | Crystal structure of the pre-cleaved Bacillus anthracis glmS ribozyme | | Descriptor: | GLMS RIBOZYME, MAGNESIUM ION, RNA (5'-R(*AP*(A2M)P*GP*CP*GP*CP*CP*AP*GP*AP*AP*CP*U)-3'), ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-12 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
4BHN
 
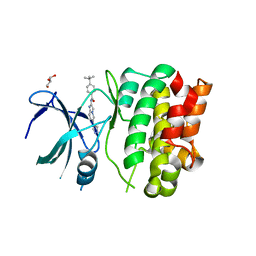 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | 4-tert-butyl-N-[6-(1H-pyrazol-4-yl)imidazo[1,2-a]pyridin-2-yl]benzamide, GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5 | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-04 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
4BIC
 
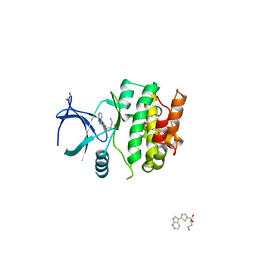 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5, N-(2-aminoethyl)-5-{1H-pyrrolo[2,3-b]pyridin-3-yl}thiophene-2-sulfonamide | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
4BIE
 
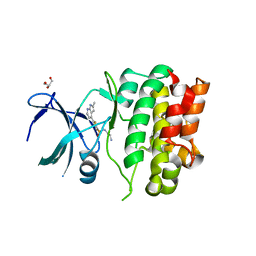 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5, N-(2-aminoethyl)-5-{2-methyl-1H-pyrrolo[2,3-b]pyridin-4-yl}thiophene-2-sulfonamide | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
4BF2
 
 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | ACETATE ION, GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5, ... | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-03-13 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
