3HV7
 
 | | Human p38 MAP Kinase in Complex with RL38 | | Descriptor: | 1-[1-(3-aminophenyl)-3-tert-butyl-1H-pyrazol-5-yl]-3-naphthalen-1-ylurea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2009-06-15 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Displacement assay for the detection of stabilizers of inactive kinase conformations.
J.Med.Chem., 53, 2010
|
|
3IW6
 
 | | Human p38 MAP Kinase in Complex with a Benzylpiperazin-Pyrrol | | Descriptor: | Mitogen-activated protein kinase 14, ethyl 4-[(4-benzylpiperazin-1-yl)carbonyl]-1-ethyl-3,5-dimethyl-1H-pyrrole-2-carboxylate, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Rauh, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-Throughput Screening To Identify Inhibitors Which Stabilize Inactive Kinase Conformations in p38alpha
J.Am.Chem.Soc., 131, 2009
|
|
3HV4
 
 | | Human p38 MAP Kinase in Complex with RL51 | | Descriptor: | 1-{3-[(6-aminoquinolin-4-yl)amino]phenyl}-3-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]urea, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mitogen-activated protein kinase 14, ... | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2009-06-15 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Displacement assay for the detection of stabilizers of inactive kinase conformations.
J.Med.Chem., 53, 2010
|
|
3GCQ
 
 | | Human P38 MAP kinase in complex with RL45 | | Descriptor: | 1-{4-[(6-aminoquinazolin-4-yl)amino]phenyl}-3-[3-tert-butyl-1-(3-methylphenyl)-1H-pyrazol-5-yl]urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2009-02-22 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of a fluorescent-tagged kinase assay system for the detection and characterization of allosteric kinase inhibitors.
J.Am.Chem.Soc., 131, 2009
|
|
3ZG4
 
 | | NMR structure of the catalytic domain from E. faecium L,D- transpeptidase | | Descriptor: | ERFK/YBIS/YCFS/YNHG | | Authors: | Lecoq, L, Dubee, V, Triboulet, S, Bougault, C, Hugonnet, J.E, Arthur, M, Simorre, J.P. | | Deposit date: | 2012-12-14 | | Release date: | 2013-04-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Structure of Enterococcus Faecium L,D---Transpeptidase Acylated by Ertapenem Provides Insight Into the Inactivation Mechanism.
Acs Chem.Biol., 8, 2013
|
|
3ZQD
 
 | | B. subtilis L,D-transpeptidase | | Descriptor: | L, D-TRANSPEPTIDASE YKUD | | Authors: | Lecoq, L, Simorre, J.-P, Bougault, C, Arthur, M, Hugonnet, J.-E, Veckerle, C, Pessey, O. | | Deposit date: | 2011-06-09 | | Release date: | 2012-05-23 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Dynamics Induced by Beta-Lactam Antibiotics in the Active Site of Bacillus subtilis L,D-Transpeptidase.
Structure, 20, 2012
|
|
3IW8
 
 | | Structure of Inactive Human p38 MAP Kinase in Complex with a Thiazole-Urea | | Descriptor: | 1-{4-[(1S)-1-amino-2-(benzyloxy)ethyl]-1,3-thiazol-2-yl}-3-(3-chloro-4-fluorophenyl)urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Rauh, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-Throughput Screening To Identify Inhibitors Which Stabilize Inactive Kinase Conformations in p38alpha
J.Am.Chem.Soc., 131, 2009
|
|
3ZGP
 
 | | NMR structure of the catalytic domain from E. faecium L,D- transpeptidase acylated by ertapenem | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, ERFK/YBIS/YCFS/YNHG | | Authors: | Lecoq, L, Triboulet, S, Dubee, V, Bougault, C, Hugonnet, J.E, Arthur, M, Simorre, J.P. | | Deposit date: | 2012-12-18 | | Release date: | 2013-04-24 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The Structure of Enterococcus Faecium L,D---Transpeptidase Acylated by Ertapenem Provides Insight Into the Inactivation Mechanism.
Acs Chem.Biol., 8, 2013
|
|
3IW7
 
 | | Human p38 MAP Kinase in Complex with an Imidazo-pyridine | | Descriptor: | 2-({4-[(4-benzylpiperidin-1-yl)carbonyl]benzyl}sulfanyl)-3H-imidazo[4,5-c]pyridine, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Rauh, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-Throughput Screening To Identify Inhibitors Which Stabilize Inactive Kinase Conformations in p38alpha
J.Am.Chem.Soc., 131, 2009
|
|
3IW5
 
 | | Human p38 MAP Kinase in Complex with an Indole Derivative | | Descriptor: | Mitogen-activated protein kinase 14, N-[2-(3-{[2-(2,3-dihydro-1,4-benzodioxin-6-ylamino)-2-oxoethyl]sulfanyl}-1H-indol-1-yl)ethyl]-3-(trifluoromethyl)benzamide, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Rauh, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High-Throughput Screening To Identify Inhibitors Which Stabilize Inactive Kinase Conformations in p38alpha
J.Am.Chem.Soc., 131, 2009
|
|
2UWJ
 
 | | Structure of the heterotrimeric complex which regulates type III secretion needle formation | | Descriptor: | NICKEL (II) ION, TYPE III EXPORT PROTEIN PSCE, TYPE III EXPORT PROTEIN PSCF, ... | | Authors: | Quinaud, M, Ple, S, Job, V, Contreras-Martel, C, Simorre, J.P, Attree, I, Dessen, A. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the heterotrimeric complex that regulates type III secretion needle formation.
Proc. Natl. Acad. Sci. U.S.A., 104, 2007
|
|
2B1W
 
 | |
1PRM
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND PLR1 (AFAPPLPRR) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
1PRL
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND PLR1 (AFAPPLPRR) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
1I5V
 
 | | SOLUTION STRUCTURE OF 2-(PYRIDO[1,2-E]PURIN-4-YL)AMINO-ETHANOL INTERCALATED IN THE DNA DUPLEX D(CGATCG)2 | | Descriptor: | 2-(PYRIDO[1,2-E]PURIN-4-YL)AMINO-ETHANOL, 5'-D(*CP*GP*AP*TP*CP*G)-3' | | Authors: | Favier, A, Blackledge, M, Simorre, J.P, Marion, D, Debousy, J.C. | | Deposit date: | 2001-03-01 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 2-(pyrido[1,2-e]purin-4-yl)amino-ethanol intercalated in the DNA duplex d(CGATCG)2.
Biochemistry, 40, 2001
|
|
1S62
 
 | | Solution structure of the Escherichia coli TolA C-terminal domain | | Descriptor: | TolA protein | | Authors: | Deprez, C, Blanchard, L, Simorre, J.-P, Gavioli, M, Guerlesquin, F, Lazdunski, C, Lloubes, R, Marion, D. | | Deposit date: | 2004-01-22 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the E.coli TolA C-terminal domain reveals conformational changes upon binding to the phage g3p N-terminal domain.
J.Mol.Biol., 346, 2005
|
|
1P6R
 
 | | Solution structure of the DNA binding domain of the repressor BlaI. | | Descriptor: | Penicillinase repressor | | Authors: | Melckebeke, H.V, Vreuls, C, Gans, P, Llabres, G, Filee, P, Joris, B, Simorre, J.P. | | Deposit date: | 2003-04-30 | | Release date: | 2003-12-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structural study of BlaI: implications for the repression of genes involved in beta-lactam antibiotic resistance.
J.Mol.Biol., 333, 2003
|
|
1RLQ
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND RLP2 (RALPPLPRY) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
1GH1
 
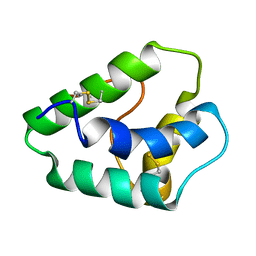 | | NMR STRUCTURES OF WHEAT NONSPECIFIC LIPID TRANSFER PROTEIN | | Descriptor: | NONSPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Gincel, E, Simorre, J.P, Caille, A, Marion, D, Ptak, M, Vovelle, F. | | Deposit date: | 2000-10-29 | | Release date: | 2000-11-22 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure in solution of a wheat lipid-transfer protein from multidimensional 1H-NMR data. A new folding for lipid carriers.
Eur.J.Biochem., 226, 1994
|
|
1K30
 
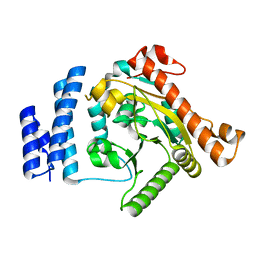 | | Crystal Structure Analysis of Squash (Cucurbita moschata) glycerol-3-phosphate (1)-acyltransferase | | Descriptor: | glycerol-3-phosphate acyltransferase | | Authors: | Turnbull, A.P, Rafferty, J.B, Sedelnikova, S.E, Slabas, A.R, Schierer, T.P, Kroon, J.T, Simon, J.W, Fawcett, T, Nishida, I, Murata, N, Rice, D.W. | | Deposit date: | 2001-10-01 | | Release date: | 2001-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analysis of the structure, substrate specificity, and mechanism of squash glycerol-3-phosphate (1)-acyltransferase.
Structure, 9, 2001
|
|
1RLP
 
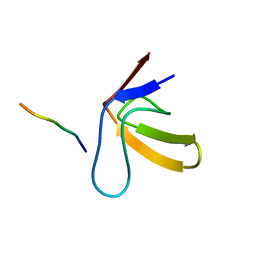 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND RLP2 (RALPPLPRY) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
2GMO
 
 | | NMR-structure of an independently folded C-terminal domain of influenza polymerase subunit PB2 | | Descriptor: | Polymerase basic protein 2 | | Authors: | Boudet, J, Tarendeau, F, Guilligay, D, Mas, P, Bougault, C.M, Cusack, S, Simorre, J.-P, Hart, D.J. | | Deposit date: | 2006-04-07 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2JLD
 
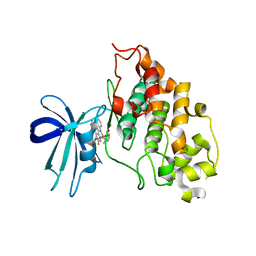 | | Extremely Tight Binding of Ruthenium Complex to Glycogen Synthase Kinase 3 | | Descriptor: | GLYCOGEN SYNTHASE KINASE-3 BETA, PEPTIDE (ALA-GLY-GLY-ALA-ALA-ALA-ALA-ALA), RUTHENIUM PYRIDOCARBAZOLE | | Authors: | Atilla-Gokcumen, G.E, Pagano, N, Streu, C, Maksimoska, J, Filippakopoulos, P, Knapp, S, Meggers, E. | | Deposit date: | 2008-09-08 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Extremely Tight Binding of a Ruthenium Complex to Glycogen Synthase Kinase 3.
Chembiochem, 9, 2008
|
|
2MHK
 
 | | E. coli LpoA N-terminal domain | | Descriptor: | Penicillin-binding protein activator LpoA | | Authors: | Jean, N.L, Bougault, C, Lodge, A, Derouaux, A, Callens, G, Egan, A, Lewis, R.J, Vollmer, W, Simorre, J. | | Deposit date: | 2013-11-26 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Elongated Structure of the Outer-Membrane Activator of Peptidoglycan Synthesis LpoA: Implications for PBP1A Stimulation.
Structure, 22, 2014
|
|
2JX0
 
 | | The paxillin-binding domain (PBD) of G Protein Coupled Receptor (GPCR)-kinase (GRK) interacting protein 1 (GIT1) | | Descriptor: | ARF GTPase-activating protein GIT1 | | Authors: | Zhang, Z, Guibao, C.D, Simmerman, J.A, Zheng, J. | | Deposit date: | 2007-11-01 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | GIT1 paxillin-binding domain is a four-helix bundle, and it binds to both paxillin LD2 and LD4 motifs.
J.Biol.Chem., 283, 2008
|
|
