1ZN8
 
 | | Human Adenine Phosphoribosyltransferase Complexed with AMP, in Space Group P1 at 1.76 A Resolution | | 分子名称: | ADENOSINE MONOPHOSPHATE, Adenine phosphoribosyltransferase, CHLORIDE ION | | 著者 | Iulek, J, Silva, M, Tomich, C.H.T.P, Thiemann, O.H. | | 登録日 | 2005-05-11 | | 公開日 | 2006-04-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structural Complexes of Human Adenine Phosphoribosyltransferase Reveal Novel Features of the APRT Catalytic Mechanism
J.Biomol.Struct.Dyn., 25, 2008
|
|
6UEK
 
 | | Structure of Urocanate Hydratase from Trypanosoma cruzi in complex with NAD+ | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Urocanate hydratase | | 著者 | Boreiko, S, Silva, M, Melo, R.F.P, Silber, A.M, Iulek, J. | | 登録日 | 2019-09-21 | | 公開日 | 2020-01-15 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Structure of Urocanate Hydratase from the protozoan Trypanosoma cruzi.
Int.J.Biol.Macromol., 146, 2019
|
|
1ZN9
 
 | | Human Adenine Phosphoribosyltransferase in Apo and AMP Complexed Forms | | 分子名称: | ADENOSINE MONOPHOSPHATE, Adenine phosphoribosyltransferase | | 著者 | Iulek, J, Silva, M, Tomich, C.H.T.P, Thiemann, O.H. | | 登録日 | 2005-05-11 | | 公開日 | 2006-04-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural Complexes of Human Adenine Phosphoribosyltransferase Reveal Novel Features of the APRT Catalytic Mechanism
J.Biomol.Struct.Dyn., 25, 2008
|
|
1S37
 
 | | Accomodation of Mispair-Aligned N3T-Ethyl-N3T DNA Interstrand Crosslink | | 分子名称: | DNA (5'-D(*CP*GP*AP*AP*AP*(TTM)P*TP*TP*TP*CP*G)-3'), DNA (5'-D(*CP*GP*AP*AP*AP*TP*TP*TP*TP*CP*G)-3') | | 著者 | da Silva, M.W, Noronha, A.M, Noll, D.M, Miller, P.S, Colvin, O.M, Gamcsik, M.P. | | 登録日 | 2004-01-12 | | 公開日 | 2005-01-25 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Accommodation of mispair aligned N3T-ethyl-N3T DNA interstrand cross link.
Biochemistry, 43, 2004
|
|
1N4B
 
 | | Solution Structure of the undecamer CGAAAC*TTTCG | | 分子名称: | 5'-D(*CP*GP*AP*AP*AP*CP*TP*TP*TP*CP*G)-3', 5'-D(*CP*GP*AP*AP*AP*D00*TP*TP*TP*CP*G)-3' | | 著者 | Webba da Silva, M, Noronha, A.M, Noll, D.M, Miller, P.S, Colvin, O.M, Gamcsik, M.P. | | 登録日 | 2002-10-30 | | 公開日 | 2003-11-04 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of a DNA Duplex Containing
Mispair-Aligned N4C-Ethyl-N4C
Interstrand Cross-Linked Cytosines
Biochemistry, 41, 2002
|
|
5J05
 
 | |
5J4P
 
 | |
5J4W
 
 | |
5J6U
 
 | |
5UR0
 
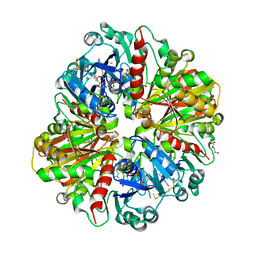 | | Crystallographic structure of glyceraldehyde-3-phosphate dehydrogenase from Naegleria gruberi | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | 著者 | Machado, A.T.P, Silva, M, Iulek, J. | | 登録日 | 2017-02-09 | | 公開日 | 2018-03-14 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Structural studies of glyceraldehyde-3-phosphate dehydrogenase from Naegleria gruberi, the first one from phylum Percolozoa.
Biochim. Biophys. Acta, 1866, 2018
|
|
2LED
 
 | |
2LEE
 
 | |
1NYD
 
 | |
2M6V
 
 | |
2MFU
 
 | |
2M6W
 
 | |
2MFT
 
 | |
3KSC
 
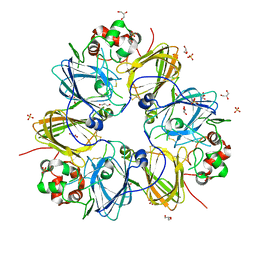 | | Crystal structure of pea prolegumin, an 11S seed globulin from Pisum sativum L. | | 分子名称: | GLYCEROL, LegA class, SULFATE ION | | 著者 | Tandang-Silvas, M.R.G, Fukuda, T, Fukuda, C, Prak, K, Cabanos, C, Kimura, A, Itoh, T, Mikami, B, Maruyama, N, Utsumi, S. | | 登録日 | 2009-11-21 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.606 Å) | | 主引用文献 | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 1804, 2010
|
|
3KGL
 
 | | Crystal structure of procruciferin, 11S globulin from Brassica napus | | 分子名称: | Cruciferin, GLYCEROL, SULFATE ION | | 著者 | Tandang-Silvas, M.R, Mikami, B, Maruyama, N, Utsumi, S. | | 登録日 | 2009-10-29 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.981 Å) | | 主引用文献 | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 1804, 2010
|
|
3QAC
 
 | | Structure of amaranth 11S proglobulin seed storage protein from Amaranthus hypochondriacus L. | | 分子名称: | 11S globulin seed storage protein | | 著者 | Tandang-Silvas, M.R, Carrazco-Pena, L, Barba de la Rosa, A.P, Osuna-Castro, J.A, Utsumi, S, Mikami, B, Maruyama, N. | | 登録日 | 2011-01-10 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.275 Å) | | 主引用文献 | Structure of amaranth 11S proglobulin, a major seed storage protein from Amaranthus hypochondriacus L.
To be Published
|
|
2XCH
 
 | | Crystal structure of PDK1 in complex with a pyrazoloquinazoline inhibitor | | 分子名称: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE 1, 8-(CYCLOHEXA-2,5-DIEN-1-YLIDENEAMINO)-1-(PIPERIDIN-4-YLMETHYL)-4,5-DIHYDRO-1H-PYRAZOLO[4,3-H]QUINAZOLINE-3-CARBOXAMIDE, GLYCEROL, ... | | 著者 | Angiolini, M, Banfi, P, Casale, E, Casuscelli, F, Fiorelli, C, Saccardo, M.B, Silvagni, M, Zuccotto, F. | | 登録日 | 2010-04-23 | | 公開日 | 2010-07-28 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-Based Optimization of Potent Pdk1 Inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XCK
 
 | | Crystal structure of PDK1 in complex with a pyrazoloquinazoline inhibitor | | 分子名称: | 1-METHYL-8-{[4-(4-METHYLPIPERAZIN-1-YL)PHENYL]AMINO}-N-[(2-METHYLPYRIDIN-4-YL)METHYL]-4,5-DIHYDRO-1H-PYRAZOLO[4,3-H]QUINAZOLINE-3-CARBOXAMIDE, 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE 1, GLYCEROL, ... | | 著者 | Angiolini, M, Banfi, P, Casale, E, Casuscelli, F, Fiorelli, C, Saccardo, M.B, Silvagni, M, Zuccotto, F. | | 登録日 | 2010-04-23 | | 公開日 | 2010-07-28 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-Based Optimization of Potent Pdk1 Inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
