6NTD
 
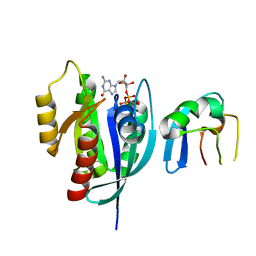 | | Crystal Structure of G12V HRas-GppNHp bound in complex with the engineered RBD variant 12 of CRAF Kinase protein | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Maisonneuve, P, Kurinov, I, Wiechmann, S, Ernst, A, Sicheri, F. | | Deposit date: | 2019-01-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Conformation-specific inhibitors of activated Ras GTPases reveal limited Ras dependency of patient-derived cancer organoids.
J.Biol.Chem., 295, 2020
|
|
5TWH
 
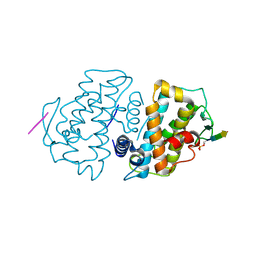 | |
1B0X
 
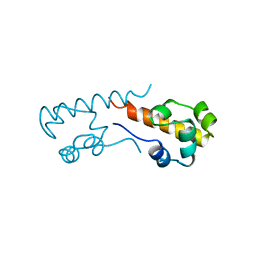 | |
3LJ1
 
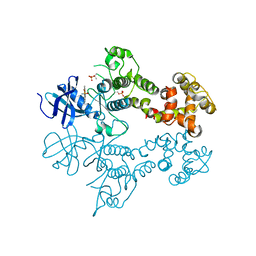 | | IRE1 complexed with Cdk1/2 Inhibitor III | | Descriptor: | 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Lee, K.P.K, Sicheri, F. | | Deposit date: | 2010-01-25 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1.
Mol.Cell, 38, 2010
|
|
1WFB
 
 | |
1WFA
 
 | |
1JPA
 
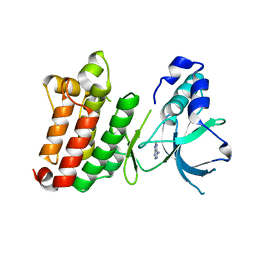 | | Crystal Structure of unphosphorylated EphB2 receptor tyrosine kinase and juxtamembrane region | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, neural kinase, Nuk=Eph/Elk/Eck family receptor-like tyrosine kinase | | Authors: | Wybenga-Groot, L.E, Pawson, T, Sicheri, F. | | Deposit date: | 2001-08-01 | | Release date: | 2001-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for autoinhibition of the Ephb2 receptor tyrosine kinase by the unphosphorylated juxtamembrane region.
Cell(Cambridge,Mass.), 106, 2001
|
|
1OPS
 
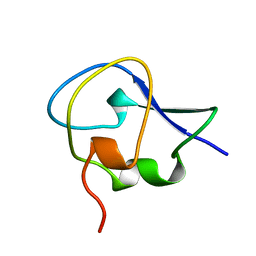 | | ICE-BINDING SURFACE ON A TYPE III ANTIFREEZE PROTEIN FROM OCEAN POUT | | Descriptor: | TYPE III ANTIFREEZE PROTEIN | | Authors: | Yang, D.S.C, Hon, W.-C, Bubanko, S, Xue, Y, Seetharaman, J, Hew, C.L, Sicheri, F. | | Deposit date: | 1997-11-17 | | Release date: | 1998-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of the ice-binding surface on a type III antifreeze protein with a "flatness function" algorithm.
Biophys.J., 74, 1998
|
|
7U3I
 
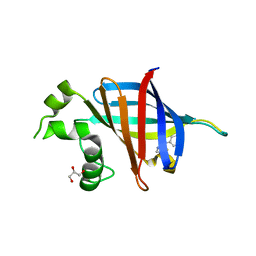 | | GID4 in complex with compound 16 | | Descriptor: | 3-{[(5S)-5-methyl-4,5-dihydro-1,3-thiazol-2-yl]amino}phenol, GLYCEROL, Glucose-induced degradation protein 4 homolog | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
7UKZ
 
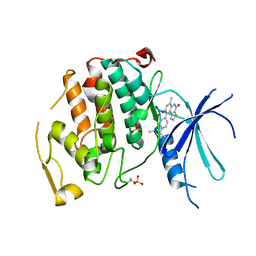 | |
7U3E
 
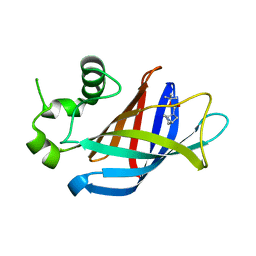 | | GID4 in complex with compound 1 | | Descriptor: | Glucose-induced degradation protein 4 homolog, tert-butyl (1S,4S)-2,5-diazabicyclo[2.2.1]heptane-2-carboxylate | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
7U3H
 
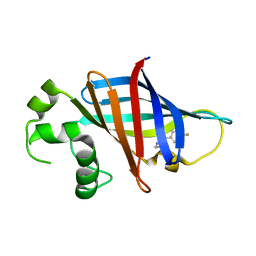 | | GID4 in complex with compound 7 | | Descriptor: | (5R)-N-(4-fluorophenyl)-5-methyl-4,5-dihydro-1,3-thiazol-2-amine, Glucose-induced degradation protein 4 homolog | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
1Q8H
 
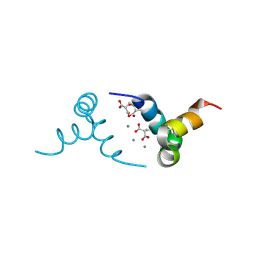 | | Crystal structure of porcine osteocalcin | | Descriptor: | CALCIUM ION, Osteocalcin | | Authors: | Hoang, Q.Q, Sicheri, F, Howard, A.J, Yang, D.S. | | Deposit date: | 2003-08-21 | | Release date: | 2003-11-11 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bone recognition mechanism of porcine osteocalcin from crystal structure.
Nature, 425, 2003
|
|
3BQ3
 
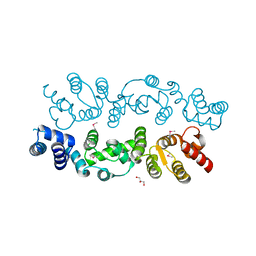 | | Crystal structure of S. cerevisiae Dcn1 | | Descriptor: | Defective in cullin neddylation protein 1, GLYCEROL | | Authors: | Chou, Y.C, Sicheri, F. | | Deposit date: | 2007-12-19 | | Release date: | 2008-01-29 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dcn1 Functions as a Scaffold-Type E3 Ligase for Cullin Neddylation.
Mol.Cell, 29, 2008
|
|
3BS7
 
 | |
1QCF
 
 | | CRYSTAL STRUCTURE OF HCK IN COMPLEX WITH A SRC FAMILY-SELECTIVE TYROSINE KINASE INHIBITOR | | Descriptor: | 1-TER-BUTYL-3-P-TOLYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, Tyrosine-protein kinase HCK | | Authors: | Schindler, T, Sicheri, F, Pico, A, Gazit, A, Levitzki, A, Kuriyan, J. | | Deposit date: | 1999-05-04 | | Release date: | 1999-06-08 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Hck in complex with a Src family-selective tyrosine kinase inhibitor.
Mol.Cell, 3, 1999
|
|
4KSL
 
 | |
4MDK
 
 | | Cdc34-ubiquitin-CC0651 complex | | Descriptor: | 4,5-dideoxy-5-(3',5'-dichlorobiphenyl-4-yl)-4-[(methoxyacetyl)amino]-L-arabinonic acid, Ubiquitin, Ubiquitin-conjugating enzyme E2 R1 | | Authors: | Ceccarelli, D.F, Orlicky, S, Tyers, M, Sicheri, F. | | Deposit date: | 2013-08-22 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6095 Å) | | Cite: | E2 enzyme inhibition by stabilization of a low-affinity interface with ubiquitin.
Nat.Chem.Biol., 10, 2014
|
|
7LUL
 
 | | Structure of the MM2 Erbin PDZ variant in complex with a high-affinity peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Teyra, J, McLaughlin, M, Ernst, A, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-02-22 | | Release date: | 2021-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Comprehensive Assessment of the Relationship Between Site -2 Specificity and Helix alpha 2 in the Erbin PDZ Domain.
J.Mol.Biol., 433, 2021
|
|
7U3G
 
 | | GID4 in complex with compound 67 | | Descriptor: | (1R)-1-phenyl-1,2,3,4-tetrahydroisoquinolin-5-amine, Glucose-induced degradation protein 4 homolog | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
7U3K
 
 | | GID4 in complex with compound 89 | | Descriptor: | GLYCEROL, Glucose-induced degradation protein 4 homolog, N-butylglycyl-4-tert-butyl-D-phenylalanyl-3-methoxy-N-methyl-L-phenylalaninamide | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
7U3J
 
 | | GID4 in complex with compound 88 | | Descriptor: | (2S)-2-{[(2S)-2-({N-[(2,4-dimethoxyphenyl)methyl]glycyl}amino)-2-(thiophen-2-yl)acetyl]amino}-N-methyl-4-phenylbutanamide, GLYCEROL, Glucose-induced degradation protein 4 homolog | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
8T9B
 
 | | Structure of the CK variant of Fab F1 (FabC-F1) in complex with the C-terminal FN3 domain of EphA2 | | Descriptor: | CK variant of Fab F1 heavy chain, CK variant of Fab F1 light chain, Ephrin type-A receptor 2 | | Authors: | Singer, A.U, Bruce, H.A, Enderle, L, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-23 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Engineered Antigen-binding Fragments for Enhanced Crystallization of Antibody:Antigen Complexes
To be Published
|
|
7M3Q
 
 | | Structure of the Smurf2 HECT Domain with a High Affinity Ubiquitin Variant (UbV) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, ... | | Authors: | Chowdhury, A, Singer, A.U, Ogunjimi, A.A, Teyra, J, Zhang, W, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-03-18 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Smurf2 HECT Domain with a High Affinity Ubiquitin Variant (UbV)
To be published
|
|
2HJN
 
 | |
