4ZOH
 
 | | Crystal structure of glyceraldehyde oxidoreductase | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Nishimasu, H, Fushinobu, S, Wakagi, T. | | Deposit date: | 2015-05-06 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Archaeal Mo-Containing Glyceraldehyde Oxidoreductase Isozymes Exhibit Diverse Substrate Specificities through Unique Subunit Assemblies.
Plos One, 11, 2016
|
|
1KXW
 
 | |
5XH7
 
 | | Crystal structure of the Acidaminococcus sp. BV3L6 Cpf1 RR variant in complex with crRNA and target DNA (TCCA PAM) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CRISPR-associated endonuclease Cpf1, ... | | Authors: | Nishimasu, H, Yamano, T, Ishitani, R, Nureki, O. | | Deposit date: | 2017-04-19 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Altered PAM Recognition by Engineered CRISPR-Cpf1
Mol. Cell, 67, 2017
|
|
5XH6
 
 | | Crystal structure of the Acidaminococcus sp. BV3L6 Cpf1 RVR variant in complex with crRNA and target DNA (TATA PAM) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CRISPR-associated endonuclease Cpf1, ... | | Authors: | Nishimasu, H, Yamano, T, Ishitani, R, Nureki, O. | | Deposit date: | 2017-04-19 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Altered PAM Recognition by Engineered CRISPR-Cpf1
Mol. Cell, 67, 2017
|
|
1O5P
 
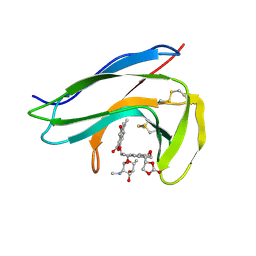 | | Solution Structure of holo-Neocarzinostatin | | Descriptor: | NEOCARZINOSTATIN-CHROMOPHORE, Neocarzinostatin | | Authors: | Takashima, H, Ishino, T, Yoshida, T, Hasuda, K, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2003-10-04 | | Release date: | 2003-10-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure Investigation for Releasing Mechanism of Neocarzinostatin Chromophore from the Holoprotein
J.Biol.Chem., 280, 2005
|
|
5CZZ
 
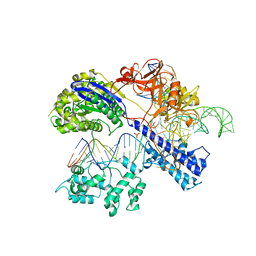 | | Crystal structure of Staphylococcus aureus Cas9 in complex with sgRNA and target DNA (TTGAAT PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, DNA (28-MER), ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2015-08-01 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Staphylococcus aureus Cas9.
Cell, 162, 2015
|
|
5AXW
 
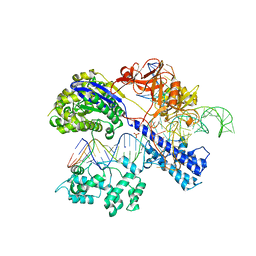 | | Crystal structure of Staphylococcus aureus Cas9 in complex with sgRNA and target DNA (TTGGGT PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, DNA (28-MER), ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2015-08-01 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Staphylococcus aureus Cas9.
Cell, 162, 2015
|
|
3WAW
 
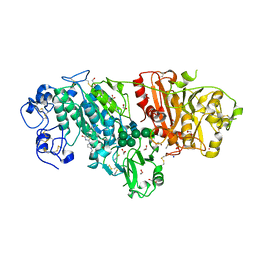 | | Crystal Structure of Autotaxin in Complex with 2BoA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-09 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Screening and X-ray Crystal Structure-based Optimization of Autotaxin (ENPP2) Inhibitors, Using a Newly Developed Fluorescence Probe
Acs Chem.Biol., 8, 2013
|
|
3WAY
 
 | | Crystal Structure of Autotaxin in Complex with 4BoA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-09 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | Screening and X-ray Crystal Structure-based Optimization of Autotaxin (ENPP2) Inhibitors, Using a Newly Developed Fluorescence Probe
Acs Chem.Biol., 8, 2013
|
|
3WAX
 
 | | Crystal Structure of Autotaxin in Complex with 3BoA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-09 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Screening and X-ray Crystal Structure-based Optimization of Autotaxin (ENPP2) Inhibitors, Using a Newly Developed Fluorescence Probe
Acs Chem.Biol., 8, 2013
|
|
3WAV
 
 | | Crystal Structure of Autotaxin in Complex with Compound 10 | | Descriptor: | (5Z)-5-(3,4-dichlorobenzylidene)-2-(4-methylpiperazin-1-yl)-1,3-thiazol-4(5H)-one, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-09 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Screening and X-ray Crystal Structure-based Optimization of Autotaxin (ENPP2) Inhibitors, Using a Newly Developed Fluorescence Probe
Acs Chem.Biol., 8, 2013
|
|
8JF5
 
 | | Crystal structure of Lysine Specific Demethylase 1 (LSD1) with TAS1440 | | Descriptor: | 4-[5-[(3~{R})-3-azanylpyrrolidin-1-yl]carbonyl-2-[2-fluoranyl-4-(2-methyl-2-oxidanyl-propyl)phenyl]phenyl]-2-fluoranyl-benzenecarbonitrile, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Fukushima, H, Machida, T, Yamashita, S, Suzuki, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | TAS1440, a histone H3 competitive LSD1 inhibitor, activates both TGF-beta and notch signaling pathways via INSM1 dissociation in neuroendocrine small cell lung cancer
To Be Published
|
|
3W1E
 
 | | Structure of FlgT, a flagellar basal body component protein | | Descriptor: | Flagella basal-body protein | | Authors: | Terashima, H, Sakuma, M, Homma, M, Imada, K. | | Deposit date: | 2012-11-14 | | Release date: | 2013-04-03 | | Last modified: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into the assembly mechanism in the supramolecular rings of the sodium-driven Vibrio flagellar motor from the structure of FlgT
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3VUX
 
 | | Crystal structure of A20 ZF7 in complex with linear ubiquitin, form II | | Descriptor: | 1,2-ETHANEDIOL, POTASSIUM ION, Polyubiquitin-C, ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2012-07-09 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-kappaB regulation
Embo J., 31, 2012
|
|
3VUW
 
 | | Crystal structure of A20 ZF7 in complex with linear ubiquitin, form I | | Descriptor: | POTASSIUM ION, Polyubiquitin-C, Tumor necrosis factor alpha-induced protein 3, ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2012-07-09 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-kappaB regulation
Embo J., 31, 2012
|
|
3VUY
 
 | | Crystal structure of A20 ZF7 in complex with linear tetraubiquitin | | Descriptor: | POTASSIUM ION, Polyubiquitin-C, Tumor necrosis factor alpha-induced protein 3, ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2012-07-09 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-kappaB regulation
Embo J., 31, 2012
|
|
1J1T
 
 | | Alginate lyase from Alteromonas sp.272 | | Descriptor: | Alginate Lyase, CALCIUM ION, SULFATE ION | | Authors: | Motoshima, H, Iwatomo, Y, Watanabe, K, Oda, T, Muramatsu, T. | | Deposit date: | 2002-12-14 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Alginate Lyase from Alteromonas sp.272
To be published
|
|
1UMG
 
 | | Crystal structure of fructose-1,6-bisphosphatase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), 385aa long conserved hypothetical protein, ... | | Authors: | Nishimasu, H, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2003-09-30 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The first crystal structure of the novel class of fructose-1,6-bisphosphatase present in thermophilic archaea.
Structure, 12, 2004
|
|
1V6R
 
 | | Solution Structure of Endothelin-1 with its C-terminal Folding | | Descriptor: | Endothelin-1 | | Authors: | Takashima, H, Mimura, N, Ohkubo, T, Yoshida, T, Tamaoki, H, Kobayashi, Y. | | Deposit date: | 2003-12-03 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Distributed Computing and NMR Constraint-Based High-Resolution Structure
Determination: Applied for Bioactive Peptide Endothelin-1 To Determine C-Terminal
Folding
J.Am.Chem.Soc., 126, 2004
|
|
1UIH
 
 | | ANALYSIS OF THE STABILIZATION OF HEN LYSOZYME WITH THE HELIX DIPOLE AND CHARGED SIDE CHAINS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSOZYME | | Authors: | Motoshima, H, Ohmura, T, Ueda, T, Imoto, T. | | Deposit date: | 1996-11-26 | | Release date: | 1997-11-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fluctuations in free or substrate-complexed lysozyme and a mutant of it detected on x-ray crystallography and comparison with those detected on NMR.
J.Biochem.(Tokyo), 131, 2002
|
|
1UIB
 
 | | ANALYSIS OF THE STABILIZATION OF HEN LYSOZYME WITH THE HELIX DIPOLE AND CHARGED SIDE CHAINS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSOZYME | | Authors: | Motoshima, H, Ohmura, T, Ueda, T, Imoto, T. | | Deposit date: | 1996-11-26 | | Release date: | 1997-11-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Fluctuations in free or substrate-complexed lysozyme and a mutant of it detected on x-ray crystallography and comparison with those detected on NMR.
J.Biochem.(Tokyo), 131, 2002
|
|
1UIF
 
 | |
1UID
 
 | |
1UIE
 
 | |
1UIC
 
 | |
