4YTM
 
 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with N-biphenyl-3-yl-2-(trifluoromethyl)benzamide | | Descriptor: | Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
4YSX
 
 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with the specific inhibitor NN23 | | Descriptor: | Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
4YTN
 
 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with N-[3-(pentafluorophenoxy)phenyl]-2-(trifluoromethyl)benzamide | | Descriptor: | Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New Insights into the Design of Inhibitors Targeted for Parasitic Anaerobic Energy Metabolism
To Be Published
|
|
4YTP
 
 | | CRYSTAL STRUCTURE OF PORCINE HEART MITOCHONDRIAL COMPLEX II BOUND WITH N-[(4-tert-butylphenyl)methyl]-2-(trifluoromethyl)benzamide | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
4YXD
 
 | | CRYSTAL STRUCTURE OF PORCINE HEART MITOCHONDRIAL COMPLEX II BOUND WITH flutolanil | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-23 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
7CII
 
 | | Crystal structure of L-methionine decarboxylase from Streptomyces sp.590 in complexed with L- methionine methyl ester (external aldimine form). | | Descriptor: | L-methionine decarboxylase, methyl (2S)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-4-methylsulfanyl-butanoate | | Authors: | Okawa, A, Shiba, T, Hayashi, M, Onoue, Y, Murota, M, Sato, D, Inagaki, J, Tamura, T, Harada, S, Inagaki, K. | | Deposit date: | 2020-07-07 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural basis for substrate specificity of l-methionine decarboxylase.
Protein Sci., 30, 2021
|
|
7E3Y
 
 | | Crystal structure of serine acetyltransferase from Salmonella typhimurium complexed with cysteine | | Descriptor: | CYSTEINE, Serine acetyltransferase | | Authors: | Momitani, K, Shiba, T, Sawa, T, Ono, K, Hurukawa, S. | | Deposit date: | 2021-02-09 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of serine acetyltransferase from Salmonella typhimurium
To Be Published
|
|
7F1P
 
 | | Crystal structure of Pseudomonas putida methionine gamma-lyase Q349S mutant ligand-free form. | | Descriptor: | L-methionine gamma-lyase | | Authors: | Okawa, A, Handa, H, Yasuda, E, Murota, M, Kudo, D, Tamura, T, Shiba, T, Inagaki, K. | | Deposit date: | 2021-06-09 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization and application of l-methionine gamma-lyase Q349S mutant enzyme with an enhanced activity toward l-homocysteine.
J.Biosci.Bioeng., 133, 2022
|
|
7F1U
 
 | | Crystal structure of Pseudomonas putida methionine gamma-lyase Q349S mutant with L-methionine intermediates | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-4-(methylsulfanyl)but-2-enoic acid, L-methionine gamma-lyase, METHIONINE | | Authors: | Okawa, A, Handa, H, Yasuda, E, Murota, M, Kudo, D, Tamura, T, Shiba, T, Inagaki, K. | | Deposit date: | 2021-06-09 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization and application of l-methionine gamma-lyase Q349S mutant enzyme with an enhanced activity toward l-homocysteine.
J.Biosci.Bioeng., 133, 2022
|
|
7F1V
 
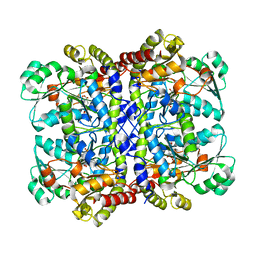 | | Crystal structure of Pseudomonas putida methionine gamma-lyase Q349S mutant with L-homocysteine intermediates | | Descriptor: | (2~{S})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-4-sulfanyl-butanoic acid, 2-AMINO-4-MERCAPTO-BUTYRIC ACID, L-methionine gamma-lyase | | Authors: | Okawa, A, Handa, H, Yasuda, E, Murota, M, Kudo, D, Tamura, T, Shiba, T, Inagaki, K. | | Deposit date: | 2021-06-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterization and application of l-methionine gamma-lyase Q349S mutant enzyme with an enhanced activity toward l-homocysteine.
J.Biosci.Bioeng., 133, 2022
|
|
7CIM
 
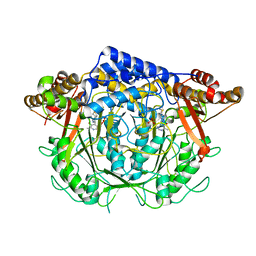 | | Crystal structure of L-methionine decarboxylase from Streptomyces sp.590 in complexed with 3-methlythiopropylamine (geminal diamine form). | | Descriptor: | L-methionine decarboxylase, [6-methyl-4-[(3-methylsulfanylpropylamino)methyl]-5-oxidanyl-pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Okawa, A, Shiba, T, Hayashi, M, Onoue, Y, Murota, M, Sato, D, Inagaki, J, Tamura, T, Harada, S, Inagaki, K. | | Deposit date: | 2020-07-07 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for substrate specificity of l-methionine decarboxylase.
Protein Sci., 30, 2021
|
|
7CIG
 
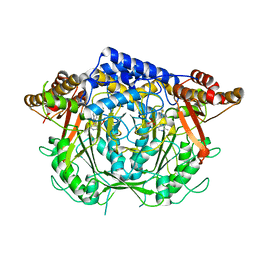 | | Crystal structure of L-methionine decarboxylase Q64A mutant from Streptomyces sp.590 in complexed with L- methionine methyl ester (geminal diamine form). | | Descriptor: | L-methionine decarboxylase, methyl (2S)-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-4-methylsulfanyl-butanoate | | Authors: | Okawa, A, Shiba, T, Hayashi, M, Onoue, Y, Murota, M, Sato, D, Inagaki, J, Tamura, T, Harada, S, Inagaki, K. | | Deposit date: | 2020-07-07 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for substrate specificity of l-methionine decarboxylase.
Protein Sci., 30, 2021
|
|
7CIJ
 
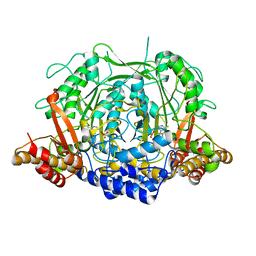 | | Crystal structure of L-methionine decarboxylase from Streptomyces sp.590 in complexed with 3-methlythiopropylamine (external aldimine form). | | Descriptor: | L-methionine decarboxylase, [6-methyl-4-[(E)-3-methylsulfanylpropyliminomethyl]-5-oxidanyl-pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Okawa, A, Shiba, T, Hayashi, M, Onoue, Y, Murota, M, Sato, D, Inagaki, J, Tamura, T, Harada, S, Inagaki, K. | | Deposit date: | 2020-07-07 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural basis for substrate specificity of l-methionine decarboxylase.
Protein Sci., 30, 2021
|
|
7CIF
 
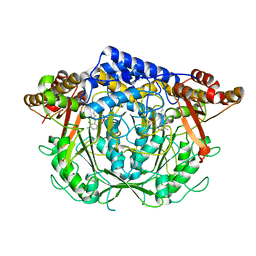 | | Crystal structure of L-methionine decarboxylase from Streptomyces sp.590 (internal aldimine form). | | Descriptor: | L-methionine decarboxylase | | Authors: | Okawa, A, Shiba, T, Hayashi, M, Onoue, Y, Murota, M, Sato, D, Inagaki, J, Tamura, T, Harada, S, Inagaki, K. | | Deposit date: | 2020-07-07 | | Release date: | 2021-01-27 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for substrate specificity of l-methionine decarboxylase.
Protein Sci., 30, 2021
|
|
1WR6
 
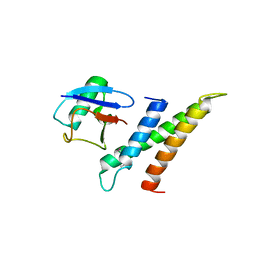 | | Crystal structure of GGA3 GAT domain in complex with ubiquitin | | Descriptor: | ADP-ribosylation factor binding protein GGA3, ubiquitin | | Authors: | Kawasaki, M, Shiba, T, Shiba, Y, Yamaguchi, Y, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Kato, K, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2004-10-12 | | Release date: | 2005-06-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism of ubiquitin recognition by GGA3 GAT domain.
Genes Cells, 10, 2005
|
|
6J9Q
 
 | | Crystal structure of Trypanosoma brucei gambiense glycerol kinase complex with AMP-PNP. | | Descriptor: | GLYCEROL, Glycerol kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Balogun, E.O, Chishima, T, Ichinose, M, Inaoka, D.K, Kido, Y, Ibrahim, B, de Koning, H, McKerrow, J.H, Watanabe, Y, Nozaki, T, Michels, P.A.M, Harada, S, Kita, K, Shiba, T. | | Deposit date: | 2019-01-24 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Reaction mechanism of the reverse reaction of African human trypanosomes glycerol kinase.
To Be Published
|
|
6J9V
 
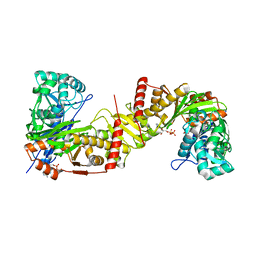 | | Crystal structure of Trypanosoma brucei gambiense glycerol kinase complex with ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Glycerol kinase | | Authors: | Balogun, E.O, Chishima, T, Ichinose, M, Inaoka, D.K, Kido, Y, Ibrahim, B, de Koning, H, McKerrow, J.H, Watanabe, Y, Nozaki, T, Michels, P.A.M, Harada, S, Kita, K, Shiba, T. | | Deposit date: | 2019-01-24 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reaction mechanism of the reverse reaction of African human trypanosomes glycerol kinase.
To Be Published
|
|
6JKR
 
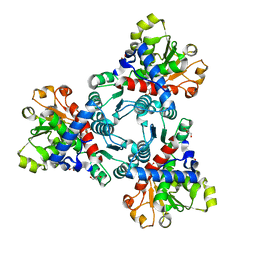 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi in complex with carbamoyl phosphate (CP) | | Descriptor: | Aspartate carbamoyltransferase, GLYCEROL, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
6JL4
 
 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi in complex with carbamoyl aspartate (CA) and phosphate (Pi) | | Descriptor: | ASPARTIC ACID, Aspartate carbamoyltransferase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-04 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
6JKS
 
 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi in complex with carbamoyl phosphate (CP) and aspartate (Asp) | | Descriptor: | ASPARTIC ACID, Aspartate carbamoyltransferase, putative, ... | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
6JL5
 
 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi in complex with aspartate (Asp) and phosphate (Pi). | | Descriptor: | ASPARTIC ACID, Aspartate carbamoyltransferase, GLYCEROL, ... | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-04 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
5AZI
 
 | | Crystal structure of glycerol kinase from Trypanosoma brucei gambiense complexed with 4NP | | Descriptor: | 4-NITROPHENYL PHOSPHATE, GLYCEROL, Glycerol kinase | | Authors: | Balogun, E.O, Inaoka, D.K, Shiba, T, Tokuoka, S.M, Tokumasu, F, Sakamoto, K, Michels, P.A.M, Harada, S, Kita, K. | | Deposit date: | 2015-10-08 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Glycerol kinase of African trypanosomes possesses an intrinsic phosphatase activity.
Biochim Biophys Acta Gen Subj, 1861, 2017
|
|
5AZJ
 
 | | Crystal structure of glycerol kinase from Trypanosoma brucei gambiense complexed with 4NP (with disulfide bridge) | | Descriptor: | 4-NITROPHENYL PHOSPHATE, GLYCEROL, Glycerol kinase | | Authors: | Balogun, E.O, Inaoka, D.K, Shiba, T, Tokuoka, S.M, Tokumasu, F, Sakamoto, K, Michels, P.A.M, Harada, S, Kita, K. | | Deposit date: | 2015-10-08 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Glycerol kinase of African trypanosomes possesses an intrinsic phosphatase activity.
Biochim Biophys Acta Gen Subj, 1861, 2017
|
|
6JKT
 
 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi in complex with N-(PHOSPHONACETYL)-L-ASPARTIC ACID (PALA). | | Descriptor: | Aspartate carbamoyltransferase, GLYCEROL, N-(PHOSPHONACETYL)-L-ASPARTIC ACID | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
6JKQ
 
 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi (Ligand-free form) | | Descriptor: | Aspartate carbamoyltransferase | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
