6A9E
 
 | | Crystal structure of the N-terminal domain of Atg2 | | Descriptor: | Endolysin,Autophagy-related protein 2 | | Authors: | Osawa, T, Noda, N.N. | | Deposit date: | 2018-07-13 | | Release date: | 2019-03-20 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Atg2 mediates direct lipid transfer between membranes for autophagosome formation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
6A9J
 
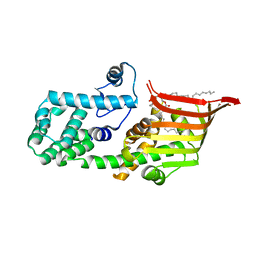 | | Crystal structure of the PE-bound N-terminal domain of Atg2 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Endolysin,Autophagy-related protein 2 | | Authors: | Osawa, T, Noda, N.N. | | Deposit date: | 2018-07-13 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Atg2 mediates direct lipid transfer between membranes for autophagosome formation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
6JP2
 
 | |
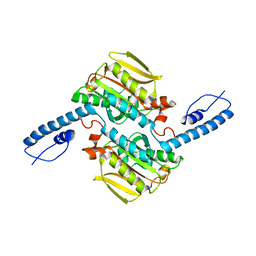
8IFJ
 
 | |
3VQV
 
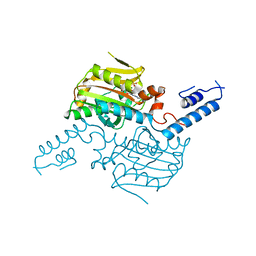 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with AMPPNP (re-refined) | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Pyrrolysine--tRNA ligase | | Authors: | Yanagisawa, T, Sumida, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-04-01 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel crystal form of pyrrolysyl-tRNA synthetase reveals the pre- and post-aminoacyl-tRNA synthesis conformational states of the adenylate and aminoacyl moieties and an asparagine residue in the catalytic site
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8GZR
 
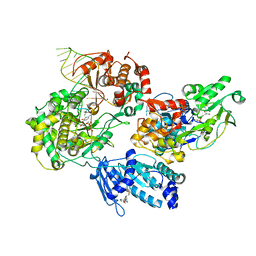 | | Cryo-EM structure of the the NS5-NS3 RNA-elongation complex | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, Genome polyprotein, MANGANESE (II) ION, ... | | Authors: | Osawa, T, Ehara, H, Sekine, S. | | Deposit date: | 2022-09-27 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of dengue virus RNA replicase complexes.
Mol.Cell, 83, 2023
|
|
8GZQ
 
 | | Cryo-EM structure of the NS5-NS3-SLA complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Genome polyprotein, RNA, ... | | Authors: | Osawa, T, Ehara, H, Sekine, S. | | Deposit date: | 2022-09-27 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of dengue virus RNA replicase complexes.
Mol.Cell, 83, 2023
|
|
8GZP
 
 | | Cryo-EM structure of the NS5-SLA complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Genome polyprotein, MAGNESIUM ION, ... | | Authors: | Osawa, T, Ehara, H, Sekine, S. | | Deposit date: | 2022-09-27 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of dengue virus RNA replicase complexes.
Mol.Cell, 83, 2023
|
|
3MGF
 
 | | Crystal Structure of Monomeric Kusabira-Orange (MKO), Orange-Emitting GFP-like Protein, at pH 7.5 | | Descriptor: | Fluorescent protein | | Authors: | Ebisawa, T, Yamamura, A, Ohtsuka, J, Kameda, Y, Hayakawa, K, Nagata, K, Tanokura, M. | | Deposit date: | 2010-04-06 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Monomeric Kusabira-Orange (MKO), Orange-Emitting GFP-like Protein, at pH 7.5
To be Published
|
|
3X1L
 
 | |
2EL7
 
 | | Crystal structure of Tryptophanyl-tRNA synthetase from Thermus thermophilus | | Descriptor: | PHOSPHATE ION, Tryptophanyl-tRNA synthetase | | Authors: | Yanagisawa, T, Sumida, T, Ishii, R, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Tryptophanyl-tRNA synthetase from Thermus thermophilus
To be published
|
|
3VQW
 
 | | Crystal structure of the SeMet substituted catalytic domain of pyrrolysyl-tRNA synthetase | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Pyrrolysine--tRNA ligase | | Authors: | Yanagisawa, T, Sumida, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-04-01 | | Release date: | 2013-01-02 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel crystal form of pyrrolysyl-tRNA synthetase reveals the pre- and post-aminoacyl-tRNA synthesis conformational states of the adenylate and aminoacyl moieties and an asparagine residue in the catalytic site
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VQY
 
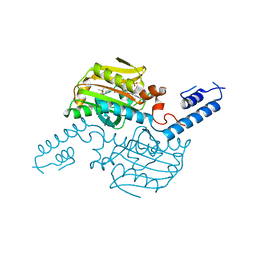 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with BocLys and AMPPNP (form 2) | | Descriptor: | MAGNESIUM ION, N~6~-(tert-butoxycarbonyl)-L-lysine, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Yanagisawa, T, Sumida, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-04-02 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel crystal form of pyrrolysyl-tRNA synthetase reveals the pre- and post-aminoacyl-tRNA synthesis conformational states of the adenylate and aminoacyl moieties and an asparagine residue in the catalytic site
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3WNC
 
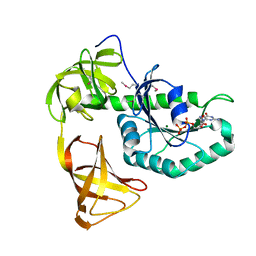 | | Crystal structure of EF-Pyl in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Protein translation elongation factor 1A | | Authors: | Yanagisawa, T, Ishii, R, Fukunaga, R, Sengoku, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2013-12-08 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A SelB/EF-Tu/aIF2 gamma-like protein from Methanosarcina mazei in the GTP-bound form binds cysteinyl-tRNA(Cys.).
J. Struct. Funct. Genomics, 16, 2015
|
|
3WNB
 
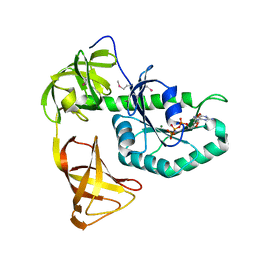 | | Crystal structure of EF-Pyl in complex with GMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Protein translation elongation factor 1A | | Authors: | Yanagisawa, T, Ishii, R, Fukunaga, R, Sengoku, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2013-12-08 | | Release date: | 2014-12-10 | | Last modified: | 2020-05-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A SelB/EF-Tu/aIF2 gamma-like protein from Methanosarcina mazei in the GTP-bound form binds cysteinyl-tRNA(Cys.).
J. Struct. Funct. Genomics, 16, 2015
|
|
3WND
 
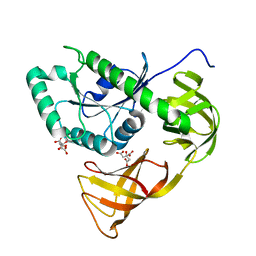 | | Crystal structure of EF-Pyl | | Descriptor: | CITRIC ACID, Protein translation elongation factor 1A | | Authors: | Yanagisawa, T, Ishii, R, Fukunaga, R, Sengoku, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2013-12-08 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A SelB/EF-Tu/aIF2 gamma-like protein from Methanosarcina mazei in the GTP-bound form binds cysteinyl-tRNA(Cys.).
J. Struct. Funct. Genomics, 16, 2015
|
|
3VQX
 
 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in triclinic crystal form | | Descriptor: | ADENOSINE MONOPHOSPHATE, PHOSPHATE ION, Pyrrolysine--tRNA ligase, ... | | Authors: | Yanagisawa, T, Sumida, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-04-02 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel crystal form of pyrrolysyl-tRNA synthetase reveals the pre- and post-aminoacyl-tRNA synthesis conformational states of the adenylate and aminoacyl moieties and an asparagine residue in the catalytic site
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2Z86
 
 | | Crystal structure of chondroitin polymerase from Escherichia coli strain K4 (K4CP) complexed with UDP-GlcUA and UDP | | Descriptor: | Chondroitin synthase, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Osawa, T, Sugiura, N, Shimada, H, Hirooka, R, Tsuji, A, Kimura, M, Kimata, K, Kakuta, Y. | | Deposit date: | 2007-09-03 | | Release date: | 2008-09-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of chondroitin polymerase from Escherichia coli K4.
Biochem. Biophys. Res. Commun., 378, 2009
|
|
2E3C
 
 | |
6ABK
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with TeocLys | | Descriptor: | (2S)-2-azanyl-6-(trimethylsilylmethoxycarbonylamino)hexanoic acid, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-22 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AAO
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with TCO*Lys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.403 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AB0
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with pAmPyLys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, N6-{[(6-aminopyridin-3-yl)methoxy]carbonyl}-L-lysine, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-19 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.441 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AAP
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with ZaeSeCys | | Descriptor: | 3-[(2-{[(benzyloxy)carbonyl]amino}ethyl)selanyl]-L-alanine, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AAD
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with mTmdZLys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, N6-[({3-[3-(trifluoromethyl)-3H-diaziren-3-yl]phenyl}methoxy)carbonyl]-L-lysine, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.444 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AAQ
 
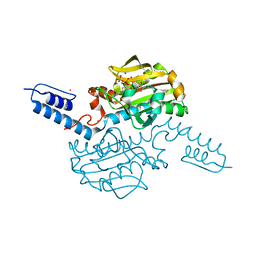 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with BCNLys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, N6-({[(1R,8S,9s)-bicyclo[6.1.0]non-4-yn-9-yl]methoxy}carbonyl)-L-lysine, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
