2NYL
 
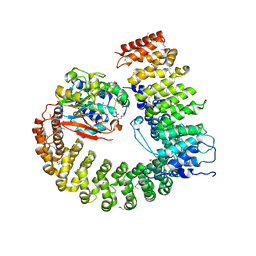 | | Crystal structure of Protein Phosphatase 2A (PP2A) holoenzyme with the catalytic subunit carboxyl terminus truncated | | Descriptor: | MANGANESE (II) ION, Protein phosphatase 2, regulatory subunit A (PR 65), ... | | Authors: | Xing, Y, Xu, Y, Chen, Y, Chao, Y, Lin, Z, Shi, Y. | | Deposit date: | 2006-11-20 | | Release date: | 2006-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the Protein Phosphatase 2A Holoenzyme.
Cell(Cambridge,Mass.), 127, 2006
|
|
2NYM
 
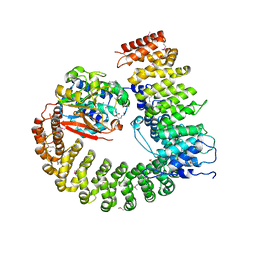 | | Crystal Structure of Protein Phosphatase 2A (PP2A) with C-terminus truncated catalytic subunit | | Descriptor: | MANGANESE (II) ION, Protein phosphatase 2, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, ... | | Authors: | Chen, Y, Xing, Y, Xu, Y, Chao, Y, Lin, Z, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2006-11-21 | | Release date: | 2006-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the Protein Phosphatase 2A Holoenzyme.
Cell(Cambridge,Mass.), 127, 2006
|
|
6ICZ
 
 | | Cryo-EM structure of a human post-catalytic spliceosome (P complex) at 3.0 angstrom | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
6J6H
 
 | | Cryo-EM structure of the yeast B*-a1 complex at an average resolution of 3.6 angstrom | | Descriptor: | ACT1 pre-mRNA, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2019-01-15 | | Release date: | 2019-04-24 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
6J6N
 
 | | Cryo-EM structure of the yeast B*-b1 complex at an average resolution of 3.86 angstrom | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2019-01-15 | | Release date: | 2019-04-24 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
2NPP
 
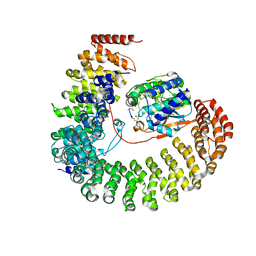 | | Structure of the Protein Phosphatase 2A Holoenzyme | | Descriptor: | MANGANESE (II) ION, Protein Phosphatase 2, regulatory subunit A (PR 65), ... | | Authors: | Xu, Y, Chen, Y, Xing, Y, Chao, Y, Shi, Y. | | Deposit date: | 2006-10-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the protein phosphatase 2A holoenzyme
Cell(Cambridge,Mass.), 127, 2006
|
|
6J6Q
 
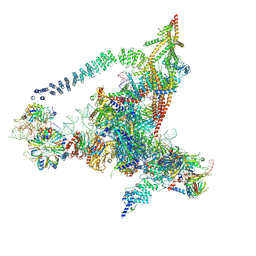 | | Cryo-EM structure of the yeast B*-b2 complex at an average resolution of 3.7 angstrom | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2019-01-15 | | Release date: | 2019-04-24 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
6KMS
 
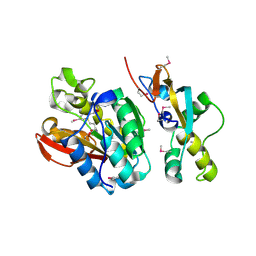 | | Crystal structure of human N6amt1-Trm112 in complex with SAM (space group I422) | | Descriptor: | Methyltransferase N6AMT1, Multifunctional methyltransferase subunit TRM112-like protein, S-ADENOSYLMETHIONINE | | Authors: | Li, W.J, Shi, Y, Zhang, T.L, Ye, J, Ding, J.P. | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-18 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insight into human N6amt1-Trm112 complex functioning as a protein methyltransferase.
Cell Discov, 5, 2019
|
|
8H6K
 
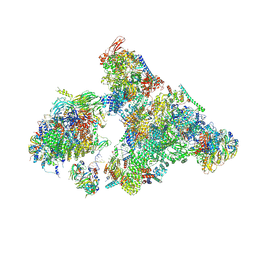 | | Cryo-EM structure of human exon-defined spliceosome in the mature B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Bai, R, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
8H6L
 
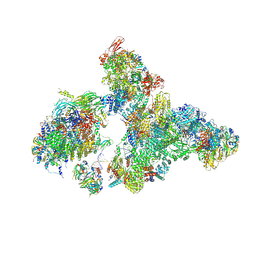 | | Cryo-EM structure of human exon-defined spliceosome in the early B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Bai, R, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
8H6J
 
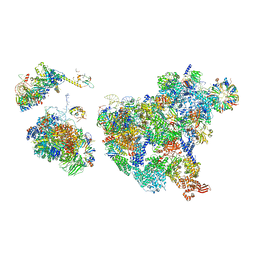 | | Cryo-EM structure of human exon-defined spliceosome in the mature pre-B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
8H6E
 
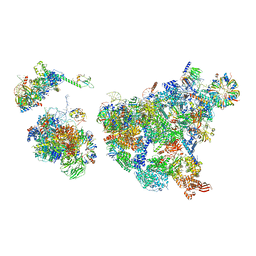 | | Cryo-EM structure of human exon-defined spliceosome in the late pre-B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Bai, R, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-17 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
2PKG
 
 | |
1HW5
 
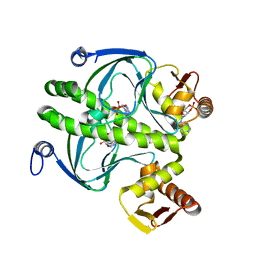 | | THE CAP/CRP VARIANT T127L/S128A | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CATABOLITE GENE ACTIVATOR | | Authors: | Chu, S.Y, Tordova, M, Gilliland, G.L, Gorshkova, I, Shi, Y. | | Deposit date: | 2001-01-09 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The structure of the T127L/S128A mutant of cAMP receptor protein facilitates promoter site binding
J.Biol.Chem., 276, 2001
|
|
1I51
 
 | | CRYSTAL STRUCTURE OF CASPASE-7 COMPLEXED WITH XIAP | | Descriptor: | CASPASE-7 SUBUNIT P11, CASPASE-7 SUBUNIT P20, X-LINKED INHIBITOR OF APOPTOSIS PROTEIN | | Authors: | Chai, J, Shi, Y. | | Deposit date: | 2001-02-23 | | Release date: | 2002-02-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of caspase-7 inhibition by XIAP.
Cell(Cambridge,Mass.), 104, 2001
|
|
1KHX
 
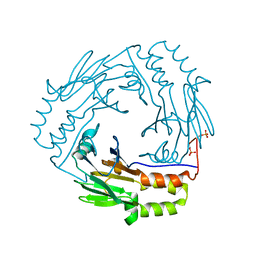 | | Crystal structure of a phosphorylated Smad2 | | Descriptor: | Smad2 | | Authors: | Wu, J.-W, Hu, M, Chai, J, Seoane, J, Huse, M, Kyin, S, Muir, T.W, Fairman, R, Massague, J, Shi, Y. | | Deposit date: | 2001-12-01 | | Release date: | 2002-02-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a phosphorylated Smad2. Recognition of phosphoserine by the MH2 domain and insights on Smad function in TGF-beta signaling.
Mol.Cell, 8, 2001
|
|
1JD5
 
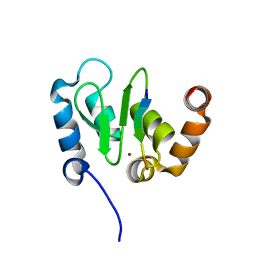 | | Crystal Structure of DIAP1-BIR2/GRIM | | Descriptor: | APOPTOSIS 1 INHIBITOR, ZINC ION, cell death protein GRIM | | Authors: | Wu, J.W, Cocina, A.E, Chai, J, Hay, B.A, Shi, Y. | | Deposit date: | 2001-06-12 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of a functional DIAP1 fragment bound to grim and hid peptides.
Mol.Cell, 8, 2001
|
|
1LCY
 
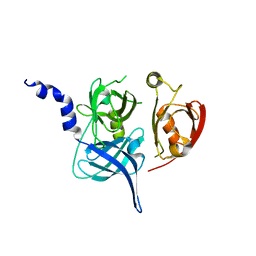 | | Crystal Structure of the Mitochondrial Serine Protease HtrA2 | | Descriptor: | HtrA2 serine protease | | Authors: | Li, W, Srinivasula, S.M, Chai, J, Li, P, Wu, J.W, Zhang, Z, Alnemri, E.S, Shi, Y. | | Deposit date: | 2002-04-07 | | Release date: | 2002-05-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the pro-apoptotic function of mitochondrial serine protease HtrA2/Omi.
Nat.Struct.Biol., 9, 2002
|
|
1JD6
 
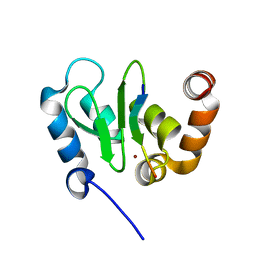 | | Crystal Structure of DIAP1-BIR2/Hid Complex | | Descriptor: | APOPTOSIS 1 INHIBITOR, ZINC ION, head involution defective protein | | Authors: | Wu, J.W, Cocina, A.E, Chai, J, Hay, B.A, Shi, Y. | | Deposit date: | 2001-06-12 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of a functional DIAP1 fragment bound to grim and hid peptides.
Mol.Cell, 8, 2001
|
|
1JD4
 
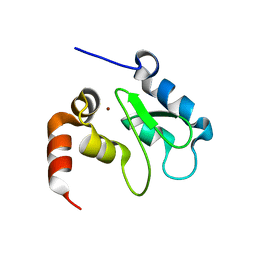 | | Crystal Structure of DIAP1-BIR2 | | Descriptor: | APOPTOSIS 1 INHIBITOR, ZINC ION | | Authors: | Wu, J.W, Cocina, A.E, Chai, J, Hay, B.A, Shi, Y. | | Deposit date: | 2001-06-12 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of a functional DIAP1 fragment bound to grim and hid peptides.
Mol.Cell, 8, 2001
|
|
2M80
 
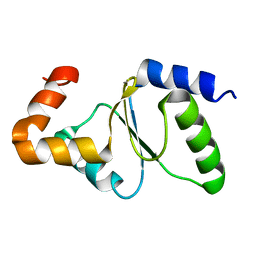 | | Solution structure of yeast dithiol glutaredoxin Grx8 | | Descriptor: | Glutaredoxin-8 | | Authors: | Tang, Y, Zhang, J, Yu, J, Wu, J, Zhou, C.Z, Shi, Y. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-guided activity enhancement and catalytic mechanism of yeast grx8
Biochemistry, 53, 2014
|
|
2MK6
 
 | |
2MOX
 
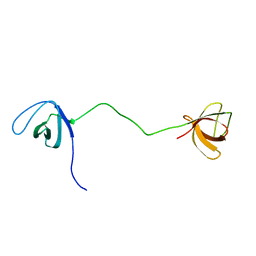 | | solution structure of tandem SH3 domain of Sorbin and SH3 domain-containing protein 1 | | Descriptor: | Sorbin and SH3 domain-containing protein 1 | | Authors: | Zhao, D, Wang, C, Zhang, J, Wu, J, Shi, Y, Zhang, Z, Gong, Q. | | Deposit date: | 2014-05-07 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural investigation of the interaction between the tandem SH3 domains of c-Cbl-associated protein and vinculin
J.Struct.Biol., 187, 2014
|
|
1M5Z
 
 | | The PDZ7 of Glutamate Receptor Interacting Protein Binds to its Target via a Novel Hydrophobic Surface Area | | Descriptor: | AMPA receptor interacting protein | | Authors: | Feng, W, Fan, J, Jiang, M, Shi, Y, Zhang, M. | | Deposit date: | 2002-07-11 | | Release date: | 2002-11-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The PDZ7 of Glutamate Receptor Interacting Protein Binds to its Target via a Novel Hydrophobic Surface Area
J.Biol.Chem., 277, 2002
|
|
4I5J
 
 | | PP2A PR70 Holoenzyme | | Descriptor: | CALCIUM ION, Serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit alpha | | Authors: | Xing, Y, Jeffery, P.D, Shi, Y. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structure of the Ca(2+)-dependent PP2A heterotrimer and insights into Cdc6 dephosphorylation.
Cell Res., 23, 2013
|
|
