1IYO
 
 | | Toho-1 beta-Lactamase In Complex With Cefotaxime | | Descriptor: | CEFOTAXIME, C3' cleaved, open, ... | | Authors: | Shimamura, T, Ibuka, A, Fushinobu, S, Wakagi, T, Ishiguro, M, Ishii, Y, Matsuzawa, H. | | Deposit date: | 2002-09-04 | | Release date: | 2002-12-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acyl-intermediate Structures of the Extended-spectrum Class A beta -Lactamase, Toho-1, in Complex with Cefotaxime, Cephalothin, and Benzylpenicillin.
J.Biol.Chem., 277, 2002
|
|
6A3L
 
 | | Crystal structure of cytochrome c' from Shewanella violacea DSS12 | | Descriptor: | Cytochrome c, HEME C | | Authors: | Suka, A, Oki, H, Kato, Y, Kawahara, K, Ohkubo, T, Maruno, T, Kobayashi, Y, Fujii, S, Wakai, S, Sambongi, Y. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-12 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Stability of cytochromes c' from psychrophilic and piezophilic Shewanella species: implications for complex multiple adaptation to low temperature and high hydrostatic pressure.
Extremophiles, 23, 2019
|
|
6AAJ
 
 | | Crystal structure of JAK2 in complex with peficitinib | | Descriptor: | 4-[[(1S,3R)-5-oxidanyl-2-adamantyl]amino]-1H-pyrrolo[2,3-b]pyridine-5-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Amano, Y, Tateishi, Y. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-15 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Discovery and structural characterization of peficitinib (ASP015K) as a novel and potent JAK inhibitor
Bioorg. Med. Chem., 26, 2018
|
|
1GDV
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME C6 FROM RED ALGA PORPHYRA YEZOENSIS AT 1.57 A RESOLUTION | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Yamada, S, Park, S.-Y, Shimizu, H, Shiro, Y, Oku, T. | | Deposit date: | 2000-10-06 | | Release date: | 2001-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure of cytochrome c6 from the red alga Porphyra yezoensis at 1. 57 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
4TWW
 
 | | Structure of SARS-3CL protease complex with a Bromobenzoyl (S,R)-N-decalin type inhibitor | | Descriptor: | (2S)-2-({[(3S,4aR,8aS)-2-(4-bromobenzoyl)decahydroisoquinolin-3-yl]methyl}amino)-3-(1H-imidazol-5-yl)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-07-02 | | Release date: | 2015-02-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors
Bioorg.Med.Chem., 23, 2015
|
|
2RVB
 
 | |
2RPV
 
 | | Solution Structure of GB1 with LBT probe | | Descriptor: | Immunoglobulin G-binding protein G, LANTHANUM (III) ION | | Authors: | Saio, T, Ogura, K, Yokochi, M, Kobashigawa, Y, Inagaki, F. | | Deposit date: | 2008-10-28 | | Release date: | 2009-09-15 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Two-point anchoring of a lanthanide-binding peptide to a target protein enhances the paramagnetic anisotropic effect
J.Biomol.Nmr, 44, 2009
|
|
1EWT
 
 | | CRYSTAL STRUCTURE OF METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 LIGAND FREE FORM I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1, SULFATE ION | | Authors: | Kunishima, N, Shimada, Y, Tsuji, Y, Jingami, H, Morikawa, K. | | Deposit date: | 2000-04-27 | | Release date: | 2000-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor.
Nature, 407, 2000
|
|
2RNF
 
 | | X-RAY CRYSTAL STRUCTURE OF HUMAN RIBONUCLEASE 4 IN COMPLEX WITH D(UP) | | Descriptor: | 2'-DEOXYURIDINE 3'-MONOPHOSPHATE, RIBONUCLEASE 4 | | Authors: | Terzyan, S.S, Peracaula, R, De Llorens, R, Tsushima, Y, Yamada, H, Seno, M, Gomis-Ruth, F.X, Coll, M. | | Deposit date: | 1998-11-03 | | Release date: | 1999-11-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure of human RNase 4, unliganded and complexed with d(Up), reveals the basis for its uridine selectivity.
J.Mol.Biol., 285, 1999
|
|
5B6Q
 
 | | Crystal structure of monomeric cytochrome c5 from Shewanella violacea | | Descriptor: | HEME C, IMIDAZOLE, Soluble cytochrome cA | | Authors: | Masanari, M, Fujii, S, Kawahara, K, Oki, H, Tsujino, H, Maruno, T, Kobayashi, Y, Ohkubo, T, Nishiyama, M, Harada, Y, Wakai, S, Sambongi, Y. | | Deposit date: | 2016-06-01 | | Release date: | 2016-10-19 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Comparative study on stabilization mechanism of monomeric cytochrome c5 from deep-sea piezophilic Shewanella violacea
Biosci.Biotechnol.Biochem., 2016
|
|
2RUK
 
 | |
2RVN
 
 | |
2RVL
 
 | |
1IZO
 
 | | Cytochrome P450 BS beta Complexed with Fatty Acid | | Descriptor: | Cytochrome P450 152A1, PALMITOLEIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, D.S, Yamada, A, Sugimoto, H, Matsunaga, I, Ogura, H, Ichihara, K, Adachi, S, Park, S.Y, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-10-10 | | Release date: | 2003-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate Recognition and Molecular Mechanism of Fatty Acid Hydroxylation by Cytochrome P450 from Bacillus subtilis. CRYSTALLOGRAPHIC, SPECTROSCOPIC, AND MUTATIONAL STUDIES.
J.Biol.Chem., 278, 2003
|
|
2STD
 
 | | SCYTALONE DEHYDRATASE COMPLEXED WITH TIGHT-BINDING INHIBITOR CARPROPAMID | | Descriptor: | ((1RS,3SR)-2,2-DICHLORO-N-[(R)-1-(4-CHLOROPHENYL)ETHYL]-1-ETHYL-3-METHYLCYCLOPROPANECARBOXAMIDE, SCYTALONE DEHYDRATASE, SULFATE ION | | Authors: | Nakasako, M, Motoyama, T, Kurahashi, Y, Yamaguchi, I. | | Deposit date: | 1997-12-21 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cryogenic X-ray crystal structure analysis for the complex of scytalone dehydratase of a rice blast fungus and its tight-binding inhibitor, carpropamid: the structural basis of tight-binding inhibition.
Biochemistry, 37, 1998
|
|
6Z1H
 
 | | Ancestral glycosidase (family 1) | | Descriptor: | ANCESTRAL RECONSTRUCTED GLYCOSIDASE, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M, Gamiz-Arco, G, Gutierrez-Rus, L, Ibarra-Molero, B, Hoshino, Y, Petrovic, D, Romero-Rivera, A, Seelig, B, Kamerlin, S.C.L, Gaucher, E.A. | | Deposit date: | 2020-05-13 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Heme-binding enables allosteric modulation in an ancient TIM-barrel glycosidase.
Nat Commun, 12, 2021
|
|
1AB9
 
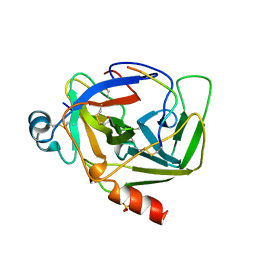 | | CRYSTAL STRUCTURE OF BOVINE GAMMA-CHYMOTRYPSIN | | Descriptor: | GAMMA-CHYMOTRYPSIN, PENTAPEPTIDE (TPGVY), SULFATE ION | | Authors: | Sugio, S, Kashima, A, Inoue, Y, Maeda, I, Nose, T, Shimohigashi, Y. | | Deposit date: | 1997-02-05 | | Release date: | 1997-08-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystal structure of a dipeptide-chymotrypsin complex in an inhibitory interaction.
Eur.J.Biochem., 255, 1998
|
|
1AFQ
 
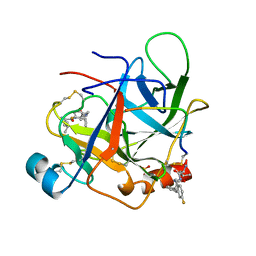 | | CRYSTAL STRUCTURE OF BOVINE GAMMA-CHYMOTRYPSIN COMPLEXED WITH A SYNTHETIC INHIBITOR | | Descriptor: | BOVINE GAMMA-CHYMOTRYPSIN, D-leucyl-N-(4-fluorobenzyl)-L-phenylalaninamide, SULFATE ION | | Authors: | Sugio, S, Kashima, A, Inoue, Y, Maeda, I, Nose, T, Shimohigashi, Y. | | Deposit date: | 1997-03-12 | | Release date: | 1997-09-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of a dipeptide-chymotrypsin complex in an inhibitory interaction.
Eur.J.Biochem., 255, 1998
|
|
1EWV
 
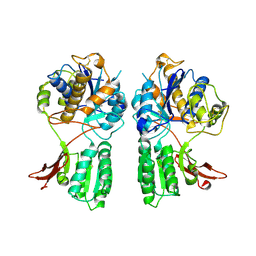 | | CRYSTAL STRUCTURE OF METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 LIGAND FREE FORM II | | Descriptor: | METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 | | Authors: | Kunishima, N, Shimada, Y, Tsuji, Y, Jingami, H, Morikawa, K. | | Deposit date: | 2000-04-27 | | Release date: | 2000-12-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor.
Nature, 407, 2000
|
|
5Z27
 
 | |
5B3I
 
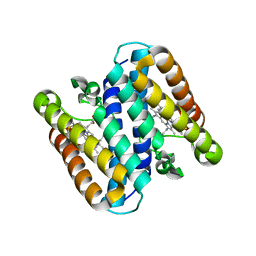 | | Homo-dimeric structure of cytochrome c' from Thermophilic Hydrogenophilus thermoluteolus | | Descriptor: | Cytochrome c prime, HEME C | | Authors: | Fujii, S, Oki, H, Kawahara, K, Yamane, D, Yamanaka, M, Maruno, T, Kobayashi, Y, Masanari, M, Wakai, S, Nishihara, H, Ohkubo, T, Sambongi, Y. | | Deposit date: | 2016-02-29 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and functional insights into thermally stable cytochrome c' from a thermophile
Protein Sci., 26, 2017
|
|
3WJ4
 
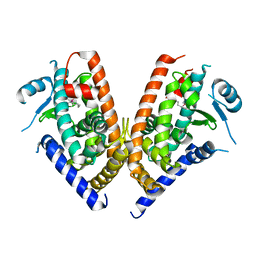 | | Crystal structure of PPARgamma ligand binding domain in complex with tributyltin | | Descriptor: | Peroxisome proliferator-activated receptor gamma, tributylstannanyl | | Authors: | Harada, S, Hiromori, Y, Fukakusa, S, Kawahara, K, Nakamura, S, Noda, M, Uchiyama, S, Fukui, K, Nishikawa, J, Nagase, H, Kobayashi, Y, Ohkubo, T, Yoshida, T, Nakanishi, T. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for PPARgamma transactivation by endocrine disrupting organotin compounds
To be Published
|
|
3WJ5
 
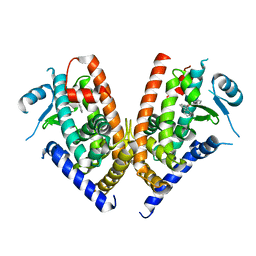 | | Crystal structure of PPARgamma ligand binding domain in complex with triphenyltin | | Descriptor: | Peroxisome proliferator-activated receptor gamma, triphenylstannanyl | | Authors: | Harada, S, Hiromori, Y, Fukakusa, S, Kawahara, K, Nakamura, S, Noda, M, Uchiyama, S, Fukui, K, Nishikawa, J, Nagase, H, Kobayashi, Y, Ohkubo, T, Yoshida, T, Nakanishi, T. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for PPARgamma transactivation by endocrine disrupting organotin compounds
To be Published
|
|
3C5V
 
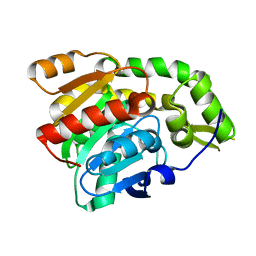 | | PP2A-specific methylesterase apo form (PME) | | Descriptor: | Protein phosphatase methylesterase 1 | | Authors: | Xing, Y, Li, Z, Chen, Y, Stock, J, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2008-02-01 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural mechanism of demethylation and inactivation of protein phosphatase 2A.
Cell(Cambridge,Mass.), 133, 2008
|
|
3C5W
 
 | | Complex between PP2A-specific methylesterase PME-1 and PP2A core enzyme | | Descriptor: | PP2A A subunit, PP2A C subunit, PP2A-specific methylesterase PME-1 | | Authors: | Xing, Y, Li, Z, Chen, Y, Stock, J, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2008-02-01 | | Release date: | 2008-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanism of demethylation and inactivation of protein phosphatase 2A.
Cell(Cambridge,Mass.), 133, 2008
|
|
