5Y2P
 
 | |
2ZXY
 
 | | Crystal Structure of Cytochrome c555 from Aquifex aeolicus | | Descriptor: | Cytochrome c552, HEME C | | Authors: | Obuchi, M, Kawahara, K, Motooka, D, Nakamura, S, Yamanaka, M, Takeda, T, Uchiyama, S, Kobayashi, Y, Ohkubo, T, Sambongi, Y. | | Deposit date: | 2009-01-09 | | Release date: | 2009-08-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Hyperstability and crystal structure of cytochrome c(555) from hyperthermophilic Aquifex aeolicus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5Y0B
 
 | | PIG GASTRIC H+,K+ - ATPASE IN COMPLEX with BYK99 | | Descriptor: | Potassium-transporting ATPase alpha chain 1, Potassium-transporting ATPase subunit beta | | Authors: | Abe, K, Shimokawa, J, Natio, M, Munson, K, Vagin, O, Sachs, G, Suzuki, H, Tani, K, Fujiyoshi, Y. | | Deposit date: | 2017-07-16 | | Release date: | 2017-08-09 | | Method: | ELECTRON CRYSTALLOGRAPHY (6.7 Å) | | Cite: | The cryo-EM structure of gastric H(+),K(+)-ATPase with bound BYK99, a high-affinity member of K(+)-competitive, imidazo[1,2-a]pyridine inhibitors
Sci Rep, 7, 2017
|
|
5YJA
 
 | |
5YLU
 
 | | Crystal structure of the gastric proton pump complexed with vonoprazan | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[5-(2-fluorophenyl)-1-pyridin-3-ylsulfonyl-pyrrol-3-yl]-~{N}-methyl-methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Abe, K, Irie, K, Nakanishi, H, Fujiyoshi, Y. | | Deposit date: | 2017-10-19 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.79988956 Å) | | Cite: | Crystal structures of the gastric proton pump
Nature, 556, 2018
|
|
5Z2B
 
 | |
5Z5C
 
 | | Crystal structure of hydrogen sulfide-producing enzyme (Fn1055) from Fusobacterium nucleatum: lysine-dimethylated form | | Descriptor: | CHLORIDE ION, Cysteine synthase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Kezuka, Y, Yoshida, Y, Nonaka, T. | | Deposit date: | 2018-01-17 | | Release date: | 2018-02-14 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural insights into the catalytic mechanism of cysteine (hydroxyl) lyase from the hydrogen sulfide-producing oral pathogen,
Biochem. J., 475, 2018
|
|
5XHJ
 
 | | Crystal Structure of P450BM3 with 5-Cyclohexylvaleroyl-L-Tryptophan | | Descriptor: | 5-cyclohexylpentanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Suzuki, K, Shoji, O, Stanfield, J.K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-04-21 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Control of stereoselectivity of benzylic hydroxylation catalysed by wild-type cytochrome P450BM3 using decoy molecules
CATALYSIS SCIENCE AND TECHNOLOGY, 7, 2017
|
|
2Z2T
 
 | |
5Z27
 
 | |
5XA3
 
 | | Crystal Structure of P450BM3 with Benzyloxycarbonyl-L-prolyl-L-phenylalanine | | Descriptor: | Bifunctional cytochrome P450/NADPH-P450 reductase, DIMETHYL SULFOXIDE, PHENYLALANINE, ... | | Authors: | Shoji, O, Yanagisawa, S, Stanfield, J.K, Suzuki, K, Kasai, C, Cong, Z, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-03-10 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Direct Hydroxylation of Benzene to Phenol by Cytochrome P450BM3 Triggered by Amino Acid Derivatives.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
2D0T
 
 | | Crystal structure of 4-phenylimidazole bound form of human indoleamine 2,3-dioxygenase | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-PHENYL-1H-IMIDAZOLE, Indoleamine 2,3-dioxygenase, ... | | Authors: | Sugimoto, H, Oda, S, Otsuki, T, Hino, T, Yoshida, T, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-08 | | Release date: | 2006-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human indoleamine 2,3-dioxygenase: catalytic mechanism of O2 incorporation by a heme-containing dioxygenase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2D0U
 
 | | Crystal structure of cyanide bound form of human indoleamine 2,3-dioxygenase | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CYANIDE ION, Indoleamine 2,3-dioxygenase, ... | | Authors: | Sugimoto, H, Oda, S, Otsuki, T, Hino, T, Yoshida, T, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-08 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of human indoleamine 2,3-dioxygenase: catalytic mechanism of O2 incorporation by a heme-containing dioxygenase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5XV8
 
 | |
1UBB
 
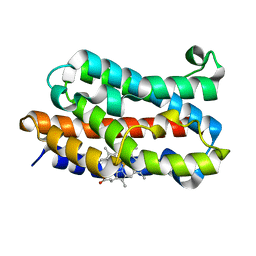 | | Crystal structure of rat HO-1 in complex with ferrous heme | | Descriptor: | Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Sakamoto, H, Higashimoto, Y, Noguchi, M, Fukuyama, K. | | Deposit date: | 2003-04-03 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Ferrous and CO-, CN(-)-, and NO-Bound Forms of Rat Heme Oxygenase-1 (HO-1) in Complex with Heme: Structural Implications for Discrimination between CO and O(2) in HO-1.
Biochemistry, 42, 2003
|
|
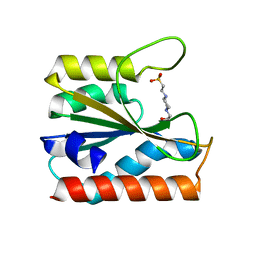
5ZBY
 
 | |
5ZC6
 
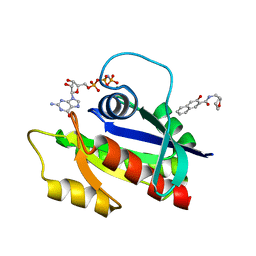 | | Solution structure of H-RasT35S mutant protein in complex with KBFM123 | | Descriptor: | 3-oxidanyl-~{N}-[[(2~{R})-oxolan-2-yl]methyl]naphthalene-2-carboxamide, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Matsumoto, S, Hayashi, Y, Hiraga, T, Matsuo, K, Kataoka, T. | | Deposit date: | 2018-02-15 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular Basis for Allosteric Inhibition of GTP-Bound H-Ras Protein by a Small-Molecule Compound Carrying a Naphthalene Ring
Biochemistry, 57, 2018
|
|
6AKF
 
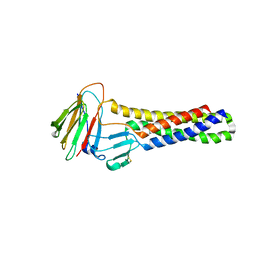 | |
2D0S
 
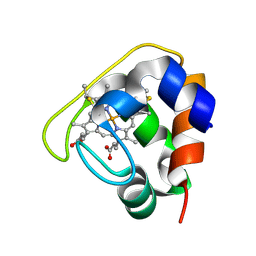 | | Crystal structure of the Cytochrome C552 from moderate thermophilic bacterium, hydrogenophilus thermoluteolus | | Descriptor: | HEME C, cytochrome c | | Authors: | Nakamura, S, Ichiki, S.I, Takashima, H, Uchiyama, S, Hasegawa, J, Kobayashi, Y, Sambongi, Y, Ohkubo, T. | | Deposit date: | 2005-08-08 | | Release date: | 2006-05-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Cytochrome c552 from a Moderate Thermophilic Bacterium, Hydrogenophilus thermoluteolus: Comparative Study on the Thermostability of Cytochrome c
Biochemistry, 45, 2006
|
|
2D4O
 
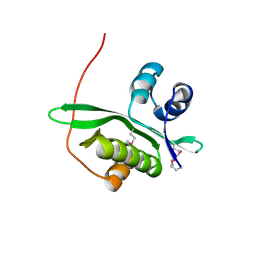 | | Crystal structure of TTHA1254 (I68M mutant) from Thermus thermophilus HB8 | | Descriptor: | hypothetical protein TTHA1254 | | Authors: | Mizohata, E, Uchikubo, T, Kinoshita, Y, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-21 | | Release date: | 2006-04-21 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of TTHA1254 (I68M mutant) from Thermus thermophilus HB8
To be Published
|
|
2E5D
 
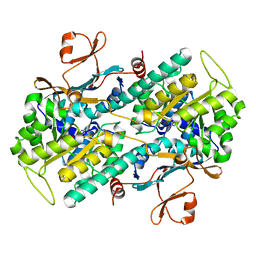 | | Crystal structure of Human NMPRTase complexed with nicotinamide | | Descriptor: | NICOTINAMIDE, Nicotinamide phosphoribosyltransferase | | Authors: | Takahashi, R, Nakamura, S, Kobayashi, Y, Ohkubo, T. | | Deposit date: | 2006-12-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and reaction mechanism of human nicotinamide phosphoribosyltransferase
J.Biochem., 147, 2010
|
|
2E5C
 
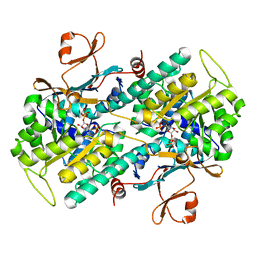 | | Crystal structure of Human NMPRTase complexed with 5'-phosphoribosyl-1'-pyrophosphate | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Nicotinamide phosphoribosyltransferase | | Authors: | Takahashi, R, Nakamura, S, Kobayashi, Y, Ohkubo, T. | | Deposit date: | 2006-12-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and reaction mechanism of human nicotinamide phosphoribosyltransferase
J.Biochem., 147, 2010
|
|
2CY4
 
 | | Crystal structure of phosphotyrosine binding (PTB) domain of epidermal growth factor receptor pathway substrate-8 (EPS8) related protein 1 from Mus musculus (form-1 crystal) | | Descriptor: | CALCIUM ION, epidermal growth factor receptor pathway substrate 8-like protein 1 | | Authors: | Mizohata, E, Hamana, H, Morita, S, Kinoshita, Y, Nagano, K, Uda, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of phosphotyrosine binding (PTB) domain of epidermal growth factor receptor pathway substrate-8 (EPS8) related protein 1 from Mus musculus (form-1 crystal)
To be Published
|
|
2D4P
 
 | | Crystal structure of TTHA1254 (wild type) from Thermus thermophilus HB8 | | Descriptor: | hypothetical protein TTHA1254 | | Authors: | Mizohata, E, Uchikubo, T, Kinoshita, Y, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-21 | | Release date: | 2006-04-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of TTHA1254 (wild type) from Thermus thermophilus HB8
To be Published
|
|
2CZ2
 
 | | Crystal structure of glutathione transferase zeta 1-1 (maleylacetoacetate isomerase) from Mus musculus (form-1 crystal) | | Descriptor: | GLUTATHIONE, GLYCEROL, Maleylacetoacetate isomerase | | Authors: | Mizohata, E, Morita, S, Kinoshita, Y, Nagano, K, Uda, H, Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-10 | | Release date: | 2006-01-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of glutathione transferase zeta 1-1 (maleylacetoacetate isomerase) from Mus musculus (form-1 crystal)
To be Published
|
|
