7XZS
 
 | | Crystal structure of Ricin A chain bound with (2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)-L-tyrosine | | Descriptor: | (2S)-2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]-3-(4-hydroxyphenyl)propanoic acid, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
7XZT
 
 | | Crystal structure of Ricin A chain bound with (2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)-D-tyrosine | | Descriptor: | (2R)-2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]-3-(4-hydroxyphenyl)propanoic acid, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
7XZW
 
 | | Crystal structure of Ricin A chain bound with (2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)-D-phenylalanine | | Descriptor: | (2R)-2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]-3-phenyl-propanoic acid, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
7XZU
 
 | | Crystal structure of Ricin A chain bound with (2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)-L-phenylalanine | | Descriptor: | (2S)-2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]-3-phenyl-propanoic acid, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
7Y02
 
 | | Crystal structure of Ricin A chain bound with (S)-2-(2-amino-4-oxo-3,4-dihydropteridine-7-carboxamido)-3-(4-fluorophenyl)propanoic acid | | Descriptor: | (2S)-2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]-3-(4-fluorophenyl)propanoic acid, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
7Y05
 
 | | Crystal structure of Ricin A chain bound with (R)-2-(2-amino-4-oxo-3,4-dihydropteridine-7-carboxamido)-2-phenylacetic acid | | Descriptor: | (2R)-2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]-2-phenyl-ethanoic acid, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
7Y06
 
 | | Crystal structure of Ricin A chain bound with (S)-2-(2-amino-4-oxo-3,4-dihydropteridine-7-carboxamido)-4-phenylbutanoic acid | | Descriptor: | (2S)-2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]-4-phenyl-butanoic acid, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
7Y03
 
 | | Crystal structure of Ricin A chain bound with (S)-2-(2-amino-4-oxo-3,4-dihydropteridine-7-carboxamido)-2-phenylacetic acid | | Descriptor: | (2S)-2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]-2-phenyl-ethanoic acid, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
7Y07
 
 | | Crystal structure of Ricin A chain bound with (S)-2-amino-N-(1-hydroxy-3-phenylpropan-2-yl)-4-oxo-3,4-dihydropteridine-7-carboxamide | | Descriptor: | 2-azanyl-4-oxidanylidene-N-[(2S)-1-oxidanyl-3-phenyl-propan-2-yl]-3H-pteridine-7-carboxamide, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
7Y08
 
 | | Crystal structure of Ricin A chain bound with (2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)glycyl-L-phenylalanine | | Descriptor: | N-[(2-amino-4-oxo-1,4-dihydropteridin-7-yl)carbonyl]glycyl-L-phenylalanine, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
3WSH
 
 | | EDTA-treated, oxidized HcgD from Methanocaldococcus jannaschii | | Descriptor: | FE (III) ION, Putative GTP cyclohydrolase 1 type 2, SULFATE ION | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2014-03-13 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A possible iron delivery function of the dinuclear iron center of HcgD in [Fe]-hydrogenase cofactor biosynthesis
Febs Lett., 588, 2014
|
|
3WVC
 
 | | Guanylylpyridinol (GP)-bound HcgF from Methanocaldococcus jannaschii | | Descriptor: | 5'-O-[(R)-hydroxy{[2-hydroxy-3,5-dimethyl-6-(2-oxo-2-sulfanylethyl)pyridin-4-yl]oxy}phosphoryl]guanosine, 5'-O-[(S)-{[2-(carboxymethyl)-6-hydroxy-3,5-dimethylpyridin-4-yl]oxy}(hydroxy)phosphoryl]guanosine, SULFATE ION, ... | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2014-05-16 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein-pyridinol thioester precursor for biosynthesis of the organometallic acyl-iron ligand in [Fe]-hydrogenase cofactor
Nat Commun, 6, 2015
|
|
3WVA
 
 | |
2CTR
 
 | | Solution structure of J-domain from human DnaJ subfamily B menber 9 | | Descriptor: | DnaJ homolog subfamily B member 9 | | Authors: | Kobayashi, N, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-24 | | Release date: | 2005-11-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of J-domain from human DnaJ subfamily B menber 9
To be Published
|
|
3WSI
 
 | | EDTA-treated, reduced HcgD from Methanocaldococcus jannaschii | | Descriptor: | FE (II) ION, PHOSPHATE ION, Putative GTP cyclohydrolase 1 type 2 | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2014-03-13 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A possible iron delivery function of the dinuclear iron center of HcgD in [Fe]-hydrogenase cofactor biosynthesis
Febs Lett., 588, 2014
|
|
3X3Y
 
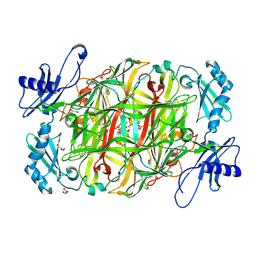 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by histamine | | Descriptor: | COPPER (II) ION, GLYCEROL, POTASSIUM ION, ... | | Authors: | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | Deposit date: | 2015-03-10 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X42
 
 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis in the presence of sodium bromide | | Descriptor: | BROMIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | Deposit date: | 2015-03-10 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.875 Å) | | Cite: | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X41
 
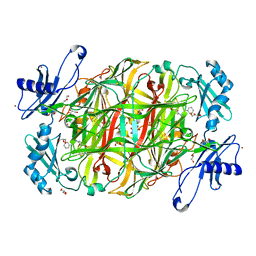 | | Copper amine oxidase from Arthrobacter globiformis: Product Schiff-base form produced by anaerobic reduction in the presence of sodium bromide | | Descriptor: | BROMIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | Deposit date: | 2015-03-10 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X3X
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by phenylethylamine | | Descriptor: | 2-PHENYL-ETHANOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | Deposit date: | 2015-03-10 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3AK4
 
 | | Crystal structure of NADH-dependent quinuclidinone reductase from agrobacterium tumefaciens | | Descriptor: | NADH-dependent quinuclidinone reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Miyakawa, T, Kataoka, M, Takeshita, D, Nomoto, F, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2010-07-07 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of NADH-dependent quinuclidinone reductase from Agrobacterium tumefaciens
To be Published
|
|
2D8B
 
 | | Solution structure of the second tandem cofilin-domain of mouse twinfilin | | Descriptor: | Twinfilin-1 | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Sato, M, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-02 | | Release date: | 2006-06-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of actin depolymerizing factor homology domains
Protein Sci., 18, 2009
|
|
2EOR
 
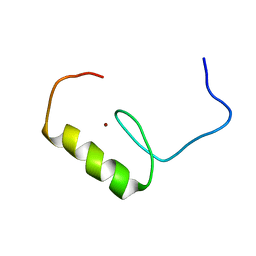 | | Solution structure of the C2H2 type zinc finger (region 255-287) of human Zinc finger protein 224 | | Descriptor: | ZINC ION, Zinc finger protein 224 | | Authors: | Tochio, N, Tomizawa, T, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-29 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 255-287) of human Zinc finger protein 224
To be Published
|
|
2EMI
 
 | | Solution structure of the C2H2 type zinc finger (region 547-579) of human Zinc finger protein 484 | | Descriptor: | ZINC ION, Zinc finger protein 484 | | Authors: | Tomizawa, T, Tochio, N, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 547-579) of human Zinc finger protein 484
To be Published
|
|
2EN2
 
 | | Solution structure of the C2H2 type zinc finger (region 598-626) of human B-cell lymphoma 6 protein | | Descriptor: | B-cell lymphoma 6 protein, ZINC ION | | Authors: | Tochio, N, Tomizawa, T, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 598-626) of human B-cell lymphoma 6 protein
To be Published
|
|
2EM4
 
 | | Solution structure of the C2H2 type zinc finger (region 724-756) of human Zinc finger protein 28 homolog | | Descriptor: | ZINC ION, Zinc finger protein 28 homolog | | Authors: | Tomizawa, T, Tochio, N, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 724-756) of human Zinc finger protein 28 homolog
To be Published
|
|
