8Y4J
 
 | |
8Y4B
 
 | |
8Y46
 
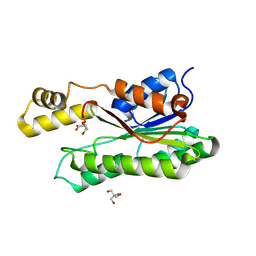 | | Crystal structure of L-2-keto-3-deoxyfuconate 4-dehydrogenase bound to L-KDF or L-2,4-DKDF | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, L-2,4-diketo-3-deoxyfuconate, ... | | Authors: | Akagashi, M, Watanabe, S. | | Deposit date: | 2024-01-30 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Crystal structure of L-2-keto-3-deoxyfuconate 4-dehydrogenase reveals a unique binding mode as a alpha-furanosyl hemiketal of substrates.
Sci Rep, 14, 2024
|
|
209D
 
 | | Structural, physical and biological characteristics of RNA:DNA binding agent N8-actinomycin D | | Descriptor: | DNA (5'-D(*GP*AP*AP*GP*CP*TP*TP*C)-3'), N8-ACTINOMYCIN D | | Authors: | Shinomiya, M, Chu, W, Carlson, R.G, Weaver, R.F, Takusagawa, F. | | Deposit date: | 1995-05-01 | | Release date: | 1995-10-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural, Physical, and Biological Characteristics of RNA.DNA Binding Agent N8-Actinomycin D.
Biochemistry, 34, 1995
|
|
7X85
 
 | |
8XWK
 
 | |
7XZZ
 
 | | Cryo-EM structure of the nucleosome in complex with p53 | | Descriptor: | Cellular tumor antigen p53, DNA (169-MER), Histone H2A type 1-B/E, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
7XZX
 
 | | Cryo-EM structure of the nucleosome in complex with p53 DNA-binding domain | | Descriptor: | Cellular tumor antigen p53, DNA (193-MER), Histone H2A type 1-B/E, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.53 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
6L88
 
 | |
2ZFD
 
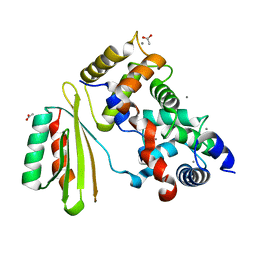 | | The crystal structure of plant specific calcium binding protein AtCBL2 in complex with the regulatory domain of AtCIPK14 | | Descriptor: | ACETIC ACID, CALCIUM ION, Calcineurin B-like protein 2, ... | | Authors: | Akaboshi, M, Hashimoto, H, Ishida, H, Koizumi, N, Sato, M, Shimizu, T. | | Deposit date: | 2007-12-29 | | Release date: | 2008-02-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of plant-specific calcium-binding protein AtCBL2 in complex with the regulatory domain of AtCIPK14
J.Mol.Biol., 377, 2008
|
|
3C2T
 
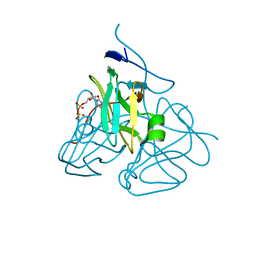 | | Evolution of chlorella virus dUTPase | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, Deoxyuridine triphosphatase, MAGNESIUM ION | | Authors: | Yamanishi, M, Homma, K, Zhang, Y, Etten, L.V.J, Moriyama, H. | | Deposit date: | 2008-01-25 | | Release date: | 2009-02-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallization and crystal-packing studies of Chlorella virus deoxyuridine triphosphatase.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
5WX8
 
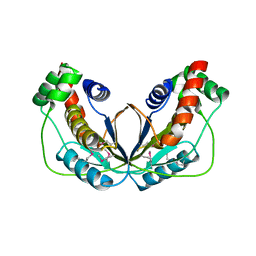 | | Human herpesvirus 6A immediate early protein 2 C-terminal domain | | Descriptor: | Immediate-early protein 2 | | Authors: | Nishimura, M, Wang, J, Wakata, A, Sakamoto, K, Mori, Y. | | Deposit date: | 2017-01-06 | | Release date: | 2017-08-02 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the DNA-Binding Domain of Human Herpesvirus 6A Immediate Early Protein 2.
J. Virol., 91, 2017
|
|
7Y00
 
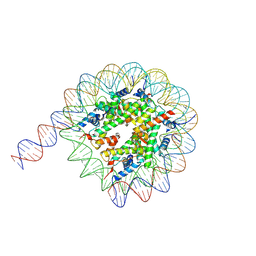 | | Cryo-EM structure of the nucleosome containing 169 base-pair DNA with a p53 target sequence | | Descriptor: | DNA (169-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
7XZY
 
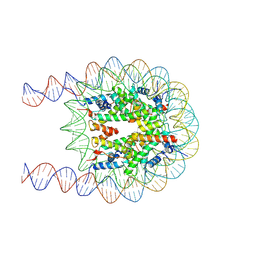 | | Cryo-EM structure of the nucleosome containing 193 base-pair DNA with a p53 target sequence | | Descriptor: | DNA (193-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
8A0K
 
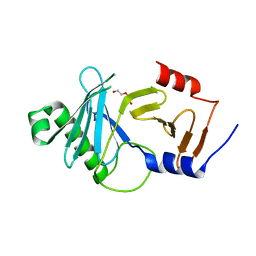 | | crystal structure of the kinetoplastid kinetochore protein Trypanosoma brucei KKT3 Divergent Polo-Box domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, Protein kinase, putative | | Authors: | Ishii, M, Ludzia, P, Marciano, G, Allen, W, Nerusheva, O.O, Akiyoshi, B. | | Deposit date: | 2022-05-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Divergent polo boxes in KKT2 bind KKT1 to initiate the kinetochore assembly cascade in Trypanosoma brucei.
Mol.Biol.Cell, 33, 2022
|
|
8A0J
 
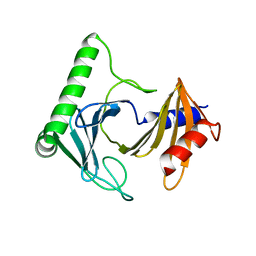 | | Crystal structure of the kinetoplastid kinetochore protein Trypanosoma congolense KKT2 divergent polo-box domain | | Descriptor: | Uncharacterized protein TCIL3000_11_11110 | | Authors: | Ishii, M, Ludzia, P, Marciano, G, Allen, W, Nerusheva, O, Akiyoshi, B. | | Deposit date: | 2022-05-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Divergent polo boxes in KKT2 bind KKT1 to initiate the kinetochore assembly cascade in Trypanosoma brucei.
Mol.Biol.Cell, 33, 2022
|
|
7OPQ
 
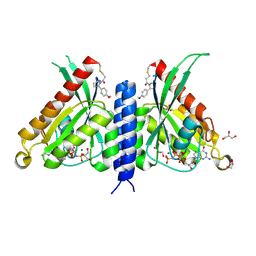 | | Rab27a fusion with Slp2a-RBDa1 effector covalent adduct with CA1 in C188 | | Descriptor: | 1-[(2~{S})-2-(4-methoxyphenyl)pyrrolidin-1-yl]propan-1-one, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Jamshidiha, M, Tersa, M, Lanyon-Hogg, T, Perez-Dorado, I, Sutherell, C.L, De Vita, E, Morgan, R.M.L, Tate, E.W, Cota, E. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Identification of the first structurally validated covalent ligands of the small GTPase RAB27A.
Rsc Med Chem, 13, 2022
|
|
7OPR
 
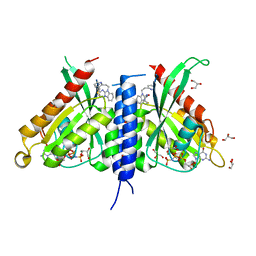 | | Rab27a fusion with Slp2a-RBDa1 effector covalent adduct with CB1 in C123 | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Jamshidiha, M, Tersa, M, Lanyon-Hogg, T, Perez-Dorado, I, Sutherell, C.L, De Vita, E, Morgan, R.M.L, Tate, E.W, Cota, E. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Identification of the first structurally validated covalent ligands of the small GTPase RAB27A.
Rsc Med Chem, 13, 2022
|
|
7OPP
 
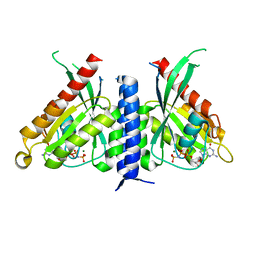 | | Crystal structure of the Rab27a fusion with Slp2a-RBDa1 effector for SF4 pocket drug targeting | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Synaptotagmin-like protein 2,Ras-related protein Rab-27A | | Authors: | Jamshidiha, M, Tersa, M, Lanyon-Hogg, T, Perez-Dorado, I, Sutherell, C.L, De Vita, E, Morgan, R.M.L, Tate, E.W, Cota, E. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Identification of the first structurally validated covalent ligands of the small GTPase RAB27A.
Rsc Med Chem, 13, 2022
|
|
6LOO
 
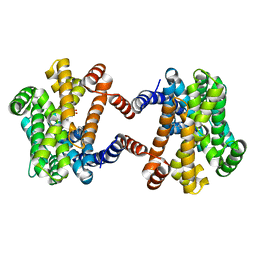 | | Crystal Structure of Class IB terpene synthase bound with geranylcitronellyl diphosphate | | Descriptor: | Tetraprenyl-beta-curcumene synthase, phosphono [(3~{R},6~{E},10~{E})-3,7,11,15-tetramethylhexadeca-6,10,14-trienyl] hydrogen phosphate, phosphono [(3~{S},6~{E},10~{E})-3,7,11,15-tetramethylhexadeca-6,10,14-trienyl] hydrogen phosphate | | Authors: | Fujihashi, M, Inagi, H, Miki, K. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Characterization of Class IB Terpene Synthase: The First Crystal Structure Bound with a Substrate Surrogate.
Acs Chem.Biol., 15, 2020
|
|
6LOP
 
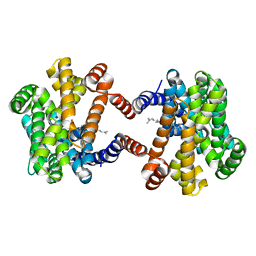 | | Crystal Structure of Class IB terpene synthase bound with geranylgeraniol | | Descriptor: | (2~{E},6~{E},10~{E})-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraen-1-ol, Tetraprenyl-beta-curcumene synthase | | Authors: | Fujihashi, M, Inagi, H, Miki, K. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Characterization of Class IB Terpene Synthase: The First Crystal Structure Bound with a Substrate Surrogate.
Acs Chem.Biol., 15, 2020
|
|
7Y7I
 
 | | chicken KNL2 in complex with the CENP-A nucleosome | | Descriptor: | Chains: I, Chains: J, Histone H2A type 1-B/E, ... | | Authors: | Ariyoshi, M, Jiang, H, Makino, F, Fukagawa, T. | | Deposit date: | 2022-06-22 | | Release date: | 2022-07-13 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | The cryo-EM structure of the CENP-A nucleosome in complex with ggKNL2.
Biorxiv, 2022
|
|
3CVF
 
 | | Crystal Structure of the carboxy terminus of Homer3 | | Descriptor: | Homer protein homolog 3 | | Authors: | Hayashi, M.K, Stearns, M.H, Giannini, V, Xu, R.-M, Sala, C, Hayashi, Y. | | Deposit date: | 2008-04-18 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The postsynaptic density proteins Homer and Shank form a polymeric network structure.
Cell(Cambridge,Mass.), 137, 2009
|
|
3CVE
 
 | | Crystal Structure of the carboxy terminus of Homer1 | | Descriptor: | Homer protein homolog 1 | | Authors: | Hayashi, M.K, Stearns, M.H, Giannini, V, Xu, R.-M, Sala, C, Hayashi, Y. | | Deposit date: | 2008-04-18 | | Release date: | 2009-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The postsynaptic density proteins Homer and Shank form a polymeric network structure.
Cell(Cambridge,Mass.), 137, 2009
|
|
2RNE
 
 | | Solution structure of the second RNA recognition motif (RRM) of TIA-1 | | Descriptor: | Tia1 protein | | Authors: | Takahashi, M, Kuwasako, K, Abe, C, Tsuda, K, Inoue, M, Terada, T, Shirouzu, M, Kobayashi, N, Kigawa, T, Taguchi, S, Guntert, P, Hayashizaki, Y, Tanaka, A, Muto, Y, Yokoyama, S. | | Deposit date: | 2007-12-19 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second RNA recognition motif (RRM) domain of murine T cell intracellular antigen-1 (TIA-1) and its RNA recognition mode
Biochemistry, 47, 2008
|
|
