4EVV
 
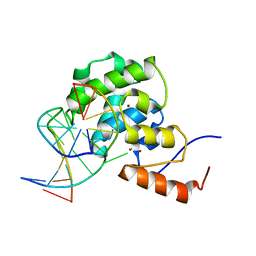 | | mouse MBD4 glycosylase domain in complex with a G:T mismatch | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*GP*TP*GP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2012-04-26 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
4EW4
 
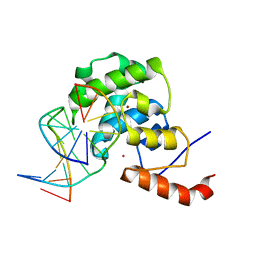 | | mouse MBD4 glycosylase domain in complex with DNA containing a ribose sugar | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*GP*(3DR)P*GP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), Methyl-CpG-binding domain protein 4, ... | | Authors: | Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2012-04-26 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.791 Å) | | Cite: | Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
2I56
 
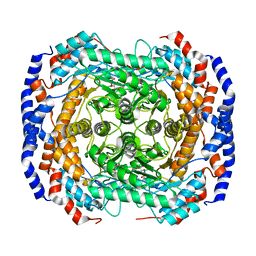 | | Crystal structure of L-Rhamnose Isomerase from Pseudomonas stutzeri with L-Rhamnose | | Descriptor: | L-RHAMNOSE, L-rhamnose isomerase, ZINC ION | | Authors: | Yoshida, H, Yamada, M, Takada, G, Izumori, K, Kamitori, S. | | Deposit date: | 2006-08-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The Structures of l-Rhamnose Isomerase from Pseudomonas stutzeri in Complexes with l-Rhamnose and d-Allose Provide Insights into Broad Substrate Specificity
J.Mol.Biol., 365, 2007
|
|
2I57
 
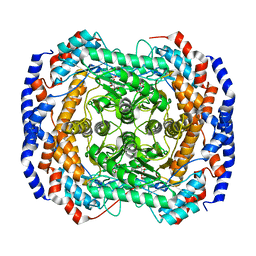 | | Crystal Structure of L-Rhamnose Isomerase from Pseudomonas stutzeri in Complex with D-Allose | | Descriptor: | D-ALLOSE, L-rhamnose isomerase, ZINC ION | | Authors: | Yoshida, H, Yamada, M, Takada, G, Izumori, K, Kamitori, S. | | Deposit date: | 2006-08-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The Structures of l-Rhamnose Isomerase from Pseudomonas stutzeri in Complexes with l-Rhamnose and d-Allose Provide Insights into Broad Substrate Specificity
J.Mol.Biol., 365, 2007
|
|
4FQZ
 
 | |
4FNC
 
 | | Human TDG in a post-reactive complex with 5-hydroxymethyluracil (5hmU) | | Descriptor: | 5-HYDROXYMETHYL URACIL, DNA (28-MER), DNA (29-MER), ... | | Authors: | Hashimoto, H, Hong, S, Bhagwat, A.S, Zhang, X, Cheng, X. | | Deposit date: | 2012-06-19 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Excision of 5-hydroxymethyluracil and 5-carboxylcytosine by the thymine DNA glycosylase domain: its structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
4EW0
 
 | | mouse MBD4 glycosylase domain in complex with a G:5hmU (5-hydroxymethyluracil) mismatch | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*GP*(5HU)P*GP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2012-04-26 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
2K3S
 
 | | HADDOCK-derived structure of the CH-domain of the smoothelin-like 1 complexed with the C-domain of apocalmodulin | | Descriptor: | Calmodulin, Smoothelin-like protein 1 | | Authors: | Ishida, H, Borman, M.A, Ostrander, J, Vogel, H.J, MacDonald, J.A. | | Deposit date: | 2008-05-15 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the calponin homology (CH) domain from the smoothelin-like 1 protein: a unique apocalmodulin-binding mode and the possible role of the C-terminal type-2 CH-domain in smooth muscle relaxation.
J.Biol.Chem., 283, 2008
|
|
2JV9
 
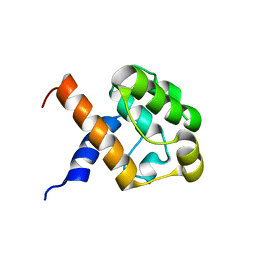 | |
2HCV
 
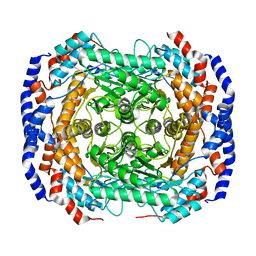 | | Crystal structure of L-rhamnose isomerase from Pseudomonas stutzeri with metal ion | | Descriptor: | L-rhamnose isomerase, ZINC ION | | Authors: | Yoshida, H, Yamada, M, Takada, G, Izumori, K, Kamitori, S. | | Deposit date: | 2006-06-19 | | Release date: | 2006-12-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structures of l-Rhamnose Isomerase from Pseudomonas stutzeri in Complexes with l-Rhamnose and d-Allose Provide Insights into Broad Substrate Specificity
J.Mol.Biol., 365, 2007
|
|
6IZZ
 
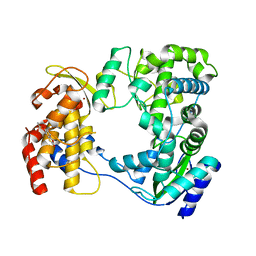 | |
6J00
 
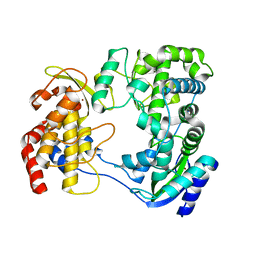 | |
7VSG
 
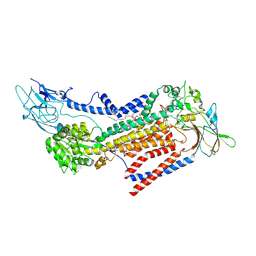 | |
7VSH
 
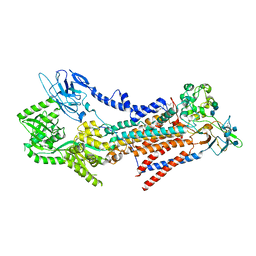 | |
1OD6
 
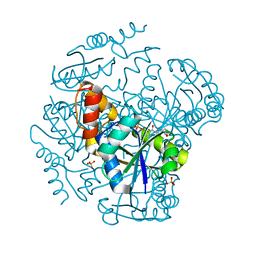 | | The Crystal Structure of Phosphopantetheine adenylyltransferase from Thermus Thermophilus in complex with 4'-phosphopantetheine | | Descriptor: | 4'-PHOSPHOPANTETHEINE, PHOSPHOPANTETHEINE ADENYLYLTRANSFERASE, SULFATE ION | | Authors: | Takahashi, H, Inagaki, E, Miyano, M, Tahirov, T.H. | | Deposit date: | 2003-02-13 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Implications for the Thermal Stability of Phosphopantetheine Adenylyltransferase from Thermus Thermophilus.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6IZX
 
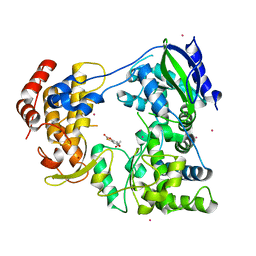 | | The RNA-dependent RNA polymerase domain of dengue 2 NS5, bound with RK-0404678 | | Descriptor: | 2-oxo-2H-1,3-benzoxathiol-5-yl acetate, COBALT (II) ION, Genome polyprotein, ... | | Authors: | Shimizu, H, Sekine, S. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of a small molecule inhibitor targeting dengue virus NS5 RNA-dependent RNA polymerase.
Plos Negl Trop Dis, 13, 2019
|
|
6IZY
 
 | |
2FAU
 
 | | Crystal structure of human vps26 | | Descriptor: | GLYCEROL, Vacuolar protein sorting 26 | | Authors: | Shi, H, Rojas, R, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2005-12-08 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The retromer subunit Vps26 has an arrestin fold and binds Vps35 through its C-terminal domain.
Nat.Struct.Mol.Biol., 13, 2006
|
|
1IV4
 
 | | Structure of 2C-Methyl-D-erythritol-2,4-cyclodiphosphate Synthase (bound form Substrate) | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CYTIDINE-5'-MONOPHOSPHATE, MAGNESIUM ION | | Authors: | Kishida, H, Wada, T, Unzai, S, Kuzuyama, T, Terada, T, Sirouzu, M, Yokoyama, S, Tame, J.R.H, Park, S.-Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-11 | | Release date: | 2002-09-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and catalytic mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate (MECDP) synthase, an enzyme in the non-mevalonate pathway of isoprenoid synthesis.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1IV3
 
 | | Structure of 2C-Methyl-D-erythritol-2,4-cyclodiphosphate Synthase (bound form MG atoms) | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, MAGNESIUM ION | | Authors: | Kishida, H, Wada, T, Unzai, S, Kuzuyama, T, Terada, T, Sirouzu, M, Yokoyama, S, Tame, J.R.H, Park, S.-Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-11 | | Release date: | 2002-09-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure and catalytic mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate (MECDP) synthase, an enzyme in the non-mevalonate pathway of isoprenoid synthesis.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1IV2
 
 | | Structure of 2C-Methyl-D-erythritol-2,4-cyclodiphosphate Synthase (bound form CDP) | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Kishida, H, Wada, T, Unzai, S, Kuzuyama, T, Terada, T, Sirouzu, M, Yokoyama, S, Tame, J.R.H, Park, S.-Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-11 | | Release date: | 2002-09-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and catalytic mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate (MECDP) synthase, an enzyme in the non-mevalonate pathway of isoprenoid synthesis.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1J1T
 
 | | Alginate lyase from Alteromonas sp.272 | | Descriptor: | Alginate Lyase, CALCIUM ION, SULFATE ION | | Authors: | Motoshima, H, Iwatomo, Y, Watanabe, K, Oda, T, Muramatsu, T. | | Deposit date: | 2002-12-14 | | Release date: | 2004-02-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Alginate Lyase from Alteromonas sp.272
To be published
|
|
1IV1
 
 | | Structure of 2C-Methyl-D-erythritol-2,4-cyclodiphosphate Synthase | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase | | Authors: | Kishida, H, Wada, T, Unzai, S, Kuzuyama, T, Terada, T, Sirouzu, M, Yokoyama, S, Tame, J.R.H, Park, S.-Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-11 | | Release date: | 2002-09-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and catalytic mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate (MECDP) synthase, an enzyme in the non-mevalonate pathway of isoprenoid synthesis.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
7X22
 
 | | Cryo-EM structure of non gastric H,K-ATPase alpha2 K794S in (2K+)E2-AlF state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Nakanishi, H, Abe, K. | | Deposit date: | 2022-02-25 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and function of H + /K + pump mutants reveal Na + /K + pump mechanisms.
Nat Commun, 13, 2022
|
|
7X21
 
 | | Cryo-EM structure of non gastric H,K-ATPase alpha2 K794A in (K+)E2-AlF state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Nakanishi, H, Abe, K. | | Deposit date: | 2022-02-25 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and function of H + /K + pump mutants reveal Na + /K + pump mechanisms.
Nat Commun, 13, 2022
|
|
