8VCT
 
 | |
5Z3K
 
 | |
1I0E
 
 | | CRYSTAL STRUCTURE OF CREATINE KINASE FROM HUMAN MUSCLE | | Descriptor: | CREATINE KINASE,M CHAIN | | Authors: | Shen, Y.-Q, Tang, L, Zhou, H.-M, Lin, Z.-J. | | Deposit date: | 2001-01-29 | | Release date: | 2003-04-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of human muscle creatine kinase.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
8VCJ
 
 | |
1LVC
 
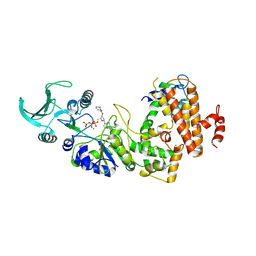 | | Crystal structure of the adenylyl cyclase domain of anthrax edema factor (EF) in complex with calmodulin and 2' deoxy, 3' anthraniloyl ATP | | Descriptor: | 3'ANTHRANILOYL-2'-DEOXY-ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, YTTERBIUM (III) ION, ... | | Authors: | Shen, Y, Lee, Y.-S, Soelaiman, S, Bergson, P, Lu, D, Chen, A, Beckingham, K, Grabarek, Z, Mrksich, M, Tang, W.-J. | | Deposit date: | 2002-05-28 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Physiological calcium concentrations regulate calmodulin binding and catalysis of adenylyl cyclase exotoxins
Embo J., 21, 2002
|
|
5C55
 
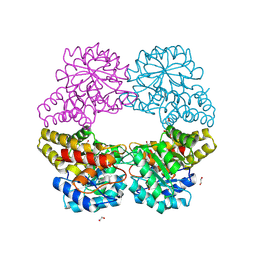 | |
1W93
 
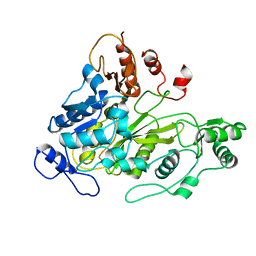 | | Crystal Structure of Biotin Carboxylase Domain of Acetyl-Coenzyme A Carboxylase from Saccharomyces cerevisiae | | Descriptor: | ACETYL-COENZYME A CARBOXYLASE | | Authors: | Shen, Y, Volrath, S.L, Weatherly, S.C, Elich, T.D, Tong, L. | | Deposit date: | 2004-10-05 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Mechanism for the Potent Inhibition of Eukaryotic Acetyl-Coenzyme a Carboxylase by Soraphen A, a Macrocyclic Polyketide Natural Product
Mol.Cell, 16, 2004
|
|
1W96
 
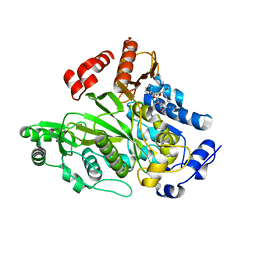 | | Crystal Structure of Biotin Carboxylase Domain of Acetyl-coenzyme A Carboxylase from Saccharomyces cerevisiae in Complex with Soraphen A | | Descriptor: | ACETYL-COENZYME A CARBOXYLASE, SORAPHEN A | | Authors: | Shen, Y, Volrath, S.L, Weatherly, S.C, Elich, T.D, Tong, L. | | Deposit date: | 2004-10-06 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Mechanism for the Potent Inhibition of Eukaryotic Acetyl-Coenzyme a Carboxylase by Soraphen A, a Macrocyclic Polyketide Natural Product
Mol.Cell, 16, 2004
|
|
5C54
 
 | |
3SUZ
 
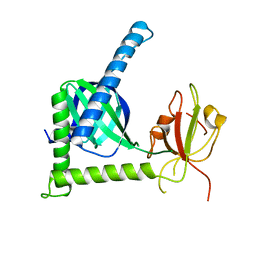 | | Crystal structure of Rat Mint2 PPC | | Descriptor: | Amyloid beta A4 precursor protein-binding family A member 2 | | Authors: | Shen, Y, Long, J, Yan, X, Xie, X. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Open-closed motion of Mint2 regulates APP metabolism
J Mol Cell Biol, 5, 2013
|
|
8GLX
 
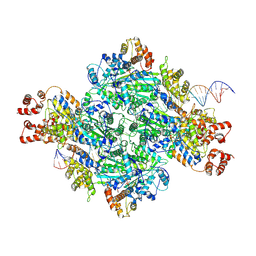 | |
6BBK
 
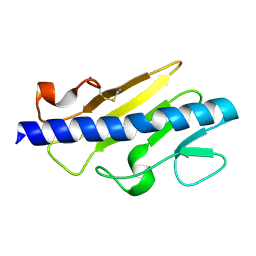 | |
3SV1
 
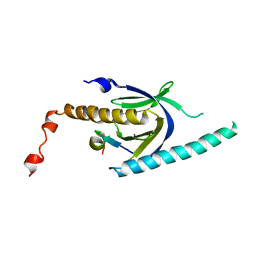 | | Crystal structure of APP peptide bound rat Mint2 PARM | | Descriptor: | Amyloid beta A4 precursor protein-binding family A member 2, Amyloid beta A4 protein | | Authors: | Shen, Y, Long, J, Yan, X, Xie, X. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Open-closed motion of Mint2 regulates APP metabolism
J Mol Cell Biol, 5, 2013
|
|
3COJ
 
 | |
3TON
 
 | |
3TOP
 
 | | Crystral Structure of the C-terminal Subunit of Human Maltase-Glucoamylase in Complex with Acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Maltase-glucoamylase, intestinal | | Authors: | Shen, Y, Qin, X.H, Ren, L.M. | | Deposit date: | 2011-09-06 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.881 Å) | | Cite: | Structural insight into substrate specificity of human intestinal maltase-glucoamylase
Protein Cell, 2, 2011
|
|
3KKR
 
 | |
1XN7
 
 | | Solution Structure of E.Coli Protein yhgG: The Northeast Structural Genomics Consortium Target ET95 | | Descriptor: | Hypothetical protein yhgG | | Authors: | Shen, Y, Acton, T, Atreya, H.S, Ma, L, Liu, G, Xiao, R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of E.Coli Protein yhgG: The Northeast Structural Genomics Consortium Target ET95
To be Published
|
|
3NKS
 
 | | Structure of human protoporphyrinogen IX oxidase | | Descriptor: | 5-[2-CHLORO-4-(TRIFLUOROMETHYL)PHENOXY]-2-NITROBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Shen, Y. | | Deposit date: | 2010-06-21 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insight into human variegate porphyria disease
Faseb J., 25, 2011
|
|
3MPX
 
 | | Crystal structure of the DH and PH-1 domains of human FGD5 | | Descriptor: | FYVE, RhoGEF and PH domain-containing protein 5, UNKNOWN ATOM OR ION | | Authors: | Shen, Y, Nedyalkova, L, Tong, Y, Tempel, W, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-27 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the DH and PH-1 domains of human FGD5
TO BE PUBLISHED
|
|
3SLU
 
 | | Crystal structure of NMB0315 | | Descriptor: | M23 peptidase domain protein, NICKEL (II) ION | | Authors: | Shen, Y, Wang, X, Yang, X, Xu, H. | | Deposit date: | 2011-06-26 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structure of outer membrane protein NMB0315 from Neisseria meningitidis.
Plos One, 6, 2011
|
|
3UB3
 
 | | D96N variant of TIR domain of Mal/TIRAP | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Shen, Y, Lin, Z. | | Deposit date: | 2011-10-23 | | Release date: | 2012-05-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Insights into TIR Domain Specificity of the Bridging Adaptor Mal in TLR4 Signaling
Plos One, 7, 2012
|
|
8GLW
 
 | |
1XFX
 
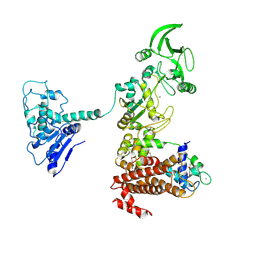 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin in the presence of 10 millimolar exogenously added calcium chloride | | Descriptor: | CALCIUM ION, Calmodulin 2, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | Deposit date: | 2004-09-15 | | Release date: | 2005-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
1XFZ
 
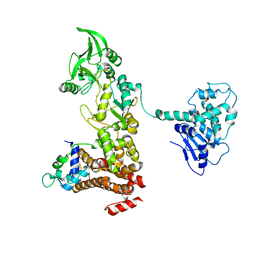 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin in the presence of 1 millimolar exogenously added calcium chloride | | Descriptor: | CALCIUM ION, Calmodulin 2, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | Deposit date: | 2004-09-15 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
