3B7J
 
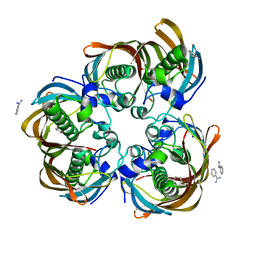 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) from Helicobacter pylori complexed with juglone | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 5-hydroxynaphthalene-1,4-dione, BENZAMIDINE, ... | | Authors: | Zhang, L, Kong, Y.H, Wu, D, Shen, X, Jiang, H.L. | | Deposit date: | 2007-10-31 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Natural product juglone targets three key enzymes from Helicobacter pylori: inhibition assay with crystal structure characterization
ACTA PHARMACOL.SIN., 29, 2008
|
|
3W4R
 
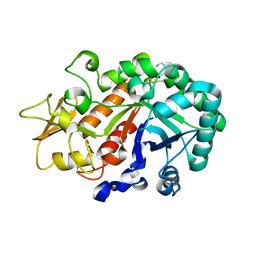 | | Crystal structure of an insect chitinase from the Asian corn borer, Ostrinia furnacalis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase | | Authors: | Chen, L, Liu, T, Zhou, Y, Shen, X, Yang, Q. | | Deposit date: | 2013-01-10 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characteristics of an insect group I chitinase, an enzyme indispensable to moulting
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WL0
 
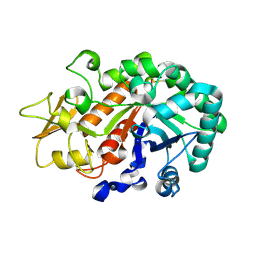 | | Crystal structure of Ostrinia furnacalis Group I chitinase catalytic domain E148A mutant in complex with a(GlcNAc)2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase | | Authors: | Chen, L, Liu, T, Zhou, Y, Chen, Q, Shen, X, Yang, Q. | | Deposit date: | 2013-11-05 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural characteristics of an insect group I chitinase, an enzyme indispensable to moulting.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WKZ
 
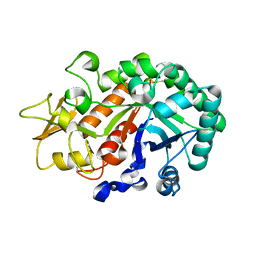 | | Crystal Structure of the Ostrinia furnacalis Group I Chitinase catalytic domain E148Q mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase | | Authors: | Chen, L, Liu, T, Zhou, Y, Chen, Q, Shen, X, Yang, Q. | | Deposit date: | 2013-11-05 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural characteristics of an insect group I chitinase, an enzyme indispensable to moulting.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WL1
 
 | | Crystal structure of Ostrinia furnacalis Group I chitinase catalytic domain in complex with reaction products (GlcNAc)2,3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Liu, T, Zhou, Y, Chen, Q, Shen, X, Yang, Q. | | Deposit date: | 2013-11-05 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | Structural characteristics of an insect group I chitinase, an enzyme indispensable to moulting.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8IRW
 
 | |
3F9G
 
 | |
3N3Y
 
 | | Crystal structure of Thymidylate Synthase X (ThyX) from Helicobacter pylori with FAD and dUMP at 2.31A resolution | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase thyX | | Authors: | Wang, K, Wang, Q, Chen, J, Chen, L, Jiang, H, Shen, X. | | Deposit date: | 2010-05-21 | | Release date: | 2011-05-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Structure, Enzymatic Characterization and Inhibitor Discovery of Thymidylate Synthase X (ThyX) from Helicobacter pylori Strain SS1
To be Published
|
|
3F9F
 
 | |
3F9H
 
 | |
3F9E
 
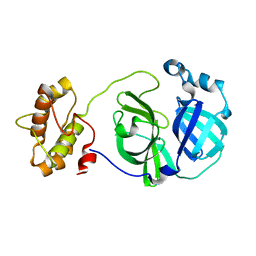 | |
3FXK
 
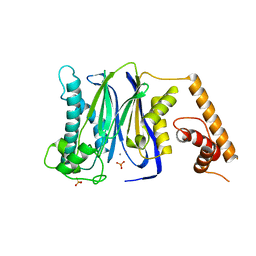 | | Crystal Structure of Human Protein phosphatase 1A (PPM1A) Bound with Phosphate at 10 mM of Mn2+ | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Protein phosphatase 1A | | Authors: | Hu, T, Wang, L, Wang, K, Jiang, H, Shen, X. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the Mn2+-dependent activation of human PPM1A
To be published
|
|
3FXJ
 
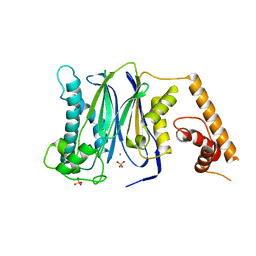 | | Crystal Structure of Human Protein phosphatase 1A (PPM1A) Bound with Phosphate at 3 mM of Mn2+ | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Protein phosphatase 1A | | Authors: | Hu, T, Wang, L, Wang, K, Jiang, H, Shen, X. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the Mn2+-dependent activation of human PPM1A
To be published
|
|
3FXL
 
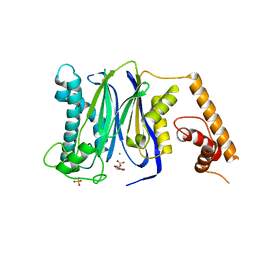 | | Crystal Structure of Human Protein phosphatase 1A (PPM1A) Bound with Citrate at 1 mM of Mn2+ | | Descriptor: | CITRATE ANION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Hu, T, Wang, L, Wang, K, Jiang, H, Shen, X. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the Mn2+-dependent activation of human PPM1A
To be published
|
|
3FXO
 
 | | Crystal Structure of Human Protein phosphatase 1A (PPM1A) Bound with Phosphate at 1 mM of Mn2+ | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Protein phosphatase 1A | | Authors: | Hu, T, Wang, L, Wang, K, Jiang, H, Shen, X. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the Mn2+-dependent activation of human PPM1A
To be published
|
|
5C4W
 
 | | Crystal structure of coxsackievirus A16 | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SODIUM ION, ... | | Authors: | Ren, J, Wang, X, Zhu, L, Hu, Z, Gao, Q, Yang, P, Li, X, Wang, J, Shen, X, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-06-18 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of Coxsackievirus A16 Capsids with Native Antigenicity: Implications for Particle Expansion, Receptor Binding, and Immunogenicity.
J.Virol., 89, 2015
|
|
5C9A
 
 | | Crystal structure of empty coxsackievirus A16 particle | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SPHINGOSINE, ... | | Authors: | Ren, J, Wang, X, Zhu, L, Hu, Z, Gao, Q, Yang, P, Li, X, Wang, J, Shen, X, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-06-26 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of Coxsackievirus A16 Capsids with Native Antigenicity: Implications for Particle Expansion, Receptor Binding, and Immunogenicity.
J.Virol., 89, 2015
|
|
1WKW
 
 | | Crystal structure of the ternary complex of eIF4E-m7GpppA-4EBP1 peptide | | Descriptor: | Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4E binding protein 1, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE | | Authors: | Tomoo, K, Matsushita, Y, Fujisaki, H, Shen, X, Miyagawa, H, Kitamura, K, Miura, K, Ishida, T. | | Deposit date: | 2004-06-10 | | Release date: | 2005-06-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for mRNA Cap-Binding regulation of eukaryotic initiation factor 4E by 4E-binding protein, studied by spectroscopic, X-ray crystal structural, and molecular dynamics simulation methods
Biochim.Biophys.Acta, 1753, 2005
|
|
5C8C
 
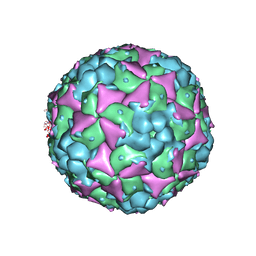 | | Crystal structure of recombinant coxsackievirus A16 capsid | | Descriptor: | CHLORIDE ION, POTASSIUM ION, STEARIC ACID, ... | | Authors: | Ren, J, Wang, X, Zhu, L, Hu, Z, Gao, Q, Yang, P, Li, X, Wang, J, Shen, X, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-06-25 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Coxsackievirus A16 Capsids with Native Antigenicity: Implications for Particle Expansion, Receptor Binding, and Immunogenicity.
J.Virol., 89, 2015
|
|
3NSM
 
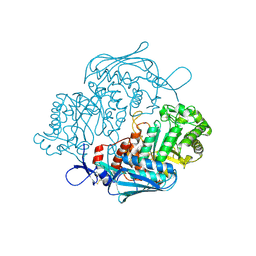 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 from Ostrinia furnacalis | | Descriptor: | N-acetylglucosaminidase | | Authors: | Zhang, H, Liu, T, Liu, F, Yang, Q, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Determinants of an Insect {beta}-N-Acetyl-D-hexosaminidase Specialized as a Chitinolytic Enzyme
J.Biol.Chem., 286, 2011
|
|
3VTR
 
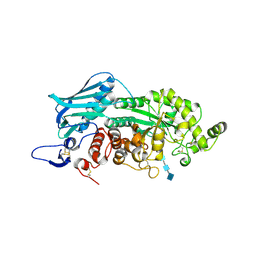 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 E328A complexed with TMG-chitotriomycin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-2-(trimethylammonio)-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetylglucosaminidase | | Authors: | Liu, T, Zhou, Y, Chen, L, Chen, W, Liu, L, Shen, X, Yang, Q. | | Deposit date: | 2012-06-02 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into cellulolytic and chitinolytic enzymes revealing crucial residues of insect beta-N-acetyl-D-hexosaminidase
Plos One, 7, 2012
|
|
3NSQ
 
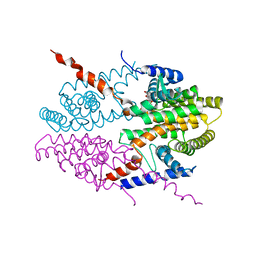 | | Crystal structure of tetrameric RXRalpha-LBD complexed with antagonist danthron | | Descriptor: | 1,8-dihydroxyanthracene-9,10-dione, Retinoid X receptor, alpha | | Authors: | Zhang, H, Hu, T, Li, L, Zhou, R, Chen, L, Hu, L, Jiang, H, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Danthron functions as a retinoic X receptor antagonist by stabilizing tetramers of the receptor.
J.Biol.Chem., 286, 2011
|
|
3NSP
 
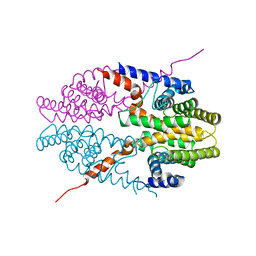 | | Crystal structure of tetrameric RXRalpha-LBD | | Descriptor: | Retinoid X receptor, alpha | | Authors: | Zhang, H, Hu, T, Li, L, Zhou, R, Chen, L, Hu, L, Jiang, H, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Danthron functions as a retinoic X receptor antagonist by stabilizing tetramers of the receptor.
J.Biol.Chem., 286, 2011
|
|
3S6T
 
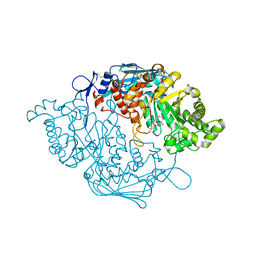 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 V327G complexed with PUGNAc | | Descriptor: | N-acetylglucosaminidase, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE | | Authors: | Zhang, H, Liu, T, Yang, Q, Shen, X. | | Deposit date: | 2011-05-26 | | Release date: | 2012-04-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Active-pocket size differentiating insectile from bacterial chitinolytic beta-N-acetyl-D-hexosaminidases.
Biochem.J., 438, 2011
|
|
3ED0
 
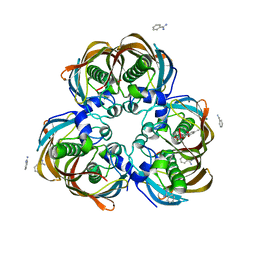 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with emodin | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, BENZAMIDINE, ... | | Authors: | Zhang, L, Zhang, H, Liu, W, Guo, Y, Shen, X, Jiang, H. | | Deposit date: | 2008-09-02 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Emodin targets the beta-hydroxyacyl-acyl carrier protein dehydratase from Helicobacter pylori: enzymatic inhibition assay with crystal structural and thermodynamic characterization
BMC MICROBIOL., 9, 2009
|
|
