7ACE
 
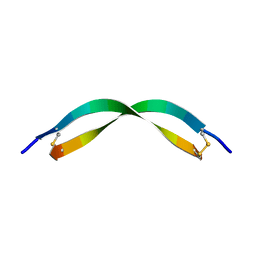 | |
5M9U
 
 | |
7ALD
 
 | |
8BWB
 
 | | Spider toxin Pha1b (PnTx3-6) from Phoneutria nigriventer targeting CaV2.x calcium channels and TRPA1 channel | | Descriptor: | Omega-ctenitoxin-Pn4a | | Authors: | Mironov, P.A, Chernaya, E.M, Paramonov, A.S, Shenkarev, Z.O. | | Deposit date: | 2022-12-06 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-05 | | Method: | SOLUTION NMR | | Cite: | Recombinant Production, NMR Solution Structure, and Membrane Interaction of the Ph alpha 1 beta Toxin, a TRPA1 Modulator from the Brazilian Armed Spider Phoneutria nigriventer .
Toxins, 15, 2023
|
|
8B4R
 
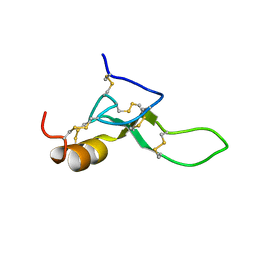
 | | Antimicrobial peptide capitellacin from polychaeta Capitella teleta in DPC (dodecylphosphocholine) micelles, monomeric form | | Descriptor: | BRICHOS domain-containing protein | | Authors: | Mironov, P.A, Reznikova, O.V, Paramonov, A.S, Shenkarev, Z.O. | | Deposit date: | 2022-09-21 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Dimerization of the beta-Hairpin Membrane-Active Cationic Antimicrobial Peptide Capitellacin from Marine Polychaeta: An NMR Structural and Thermodynamic Study.
Biomolecules, 14, 2024
|
|
8B4S
 
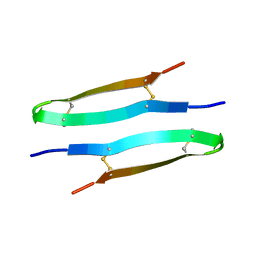 | | Antimicrobial peptide capitellacin from polychaeta Capitella teleta in DPC (dodecylphosphocholine) micelles, dimeric form | | Descriptor: | BRICHOS domain-containing protein | | Authors: | Mironov, P.A, Reznikova, O.V, Paramonov, A.S, Shenkarev, Z.O. | | Deposit date: | 2022-09-21 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Dimerization of the beta-Hairpin Membrane-Active Cationic Antimicrobial Peptide Capitellacin from Marine Polychaeta: An NMR Structural and Thermodynamic Study.
Biomolecules, 14, 2024
|
|
2MQU
 
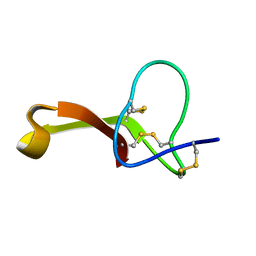 | | Spatial structure of Hm-3, a membrane-active spider toxin affecting sodium channels | | Descriptor: | Neurotoxin Hm-3 | | Authors: | Myshkin, M.Y, Shenkarev, Z.O, Paramonov, A.S, Berkut, A.A, Grishin, E.V, Vassilevski, A.A. | | Deposit date: | 2014-06-27 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of membrane-active toxin from crab spider Heriaeus melloteei suggests parallel evolution of sodium channel gating modifiers in Araneomorphae and Mygalomorphae.
J. Biol. Chem., 290, 2015
|
|
2N99
 
 | | Solution structure of the SLURP-2, a secreted isoform of Lynx1 | | Descriptor: | Ly-6/neurotoxin-like protein 1 | | Authors: | Paramonov, A.S, Shenkarev, Z.O, Lyukmanova, E.N, Arseniev, A.S. | | Deposit date: | 2015-11-11 | | Release date: | 2016-09-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Secreted Isoform of Human Lynx1 (SLURP-2): Spatial Structure and Pharmacology of Interactions with Different Types of Acetylcholine Receptors.
Sci Rep, 6, 2016
|
|
6ZZF
 
 | |
6ZSO
 
 | | Solution structure of the water-soluble LU-domain of human Lypd6b protein | | Descriptor: | Ly6/PLAUR domain-containing protein 6B | | Authors: | Tsarev, A.V, Kulbatskii, D.S, Paramonov, A.S, Lyukmanova, E.N, Shenkarev, Z.O. | | Deposit date: | 2020-07-16 | | Release date: | 2021-01-13 | | Method: | SOLUTION NMR | | Cite: | Structural Diversity and Dynamics of Human Three-Finger Proteins Acting on Nicotinic Acetylcholine Receptors.
Int J Mol Sci, 21, 2020
|
|
8F7L
 
 | | Human NAMPT in complex with substrate NAM and small molecule activator ZN-29-S | | Descriptor: | (3S)-N-[(1-benzothiophen-5-yl)methyl]-1-[(2P)-2-(3-fluoro-4-methylphenyl)-2H-pyrazolo[3,4-d]pyrimidin-4-yl]piperidine-3-carboxamide, CHLORIDE ION, NICOTINAMIDE, ... | | Authors: | Ratia, K, Xiong, R, Shen, Z, Thatcher, G.R. | | Deposit date: | 2022-11-18 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human NAMPT in complex with substrate NAM and small molecule activator ZN-29-S
To Be Published
|
|
2N81
 
 | | Solution Structure of Lipid Transfer Protein From Pea Pisum Sativum | | Descriptor: | Lipid Transfer Protein | | Authors: | Paramonov, A.S, Rumynskiy, E.I, Bogdanov, I.V, Finkina, E.I, Melnikova, D.N, Ovchinnikova, T.V, Shenkarev, Z.O, Arseniev, A.S. | | Deposit date: | 2015-09-30 | | Release date: | 2016-05-11 | | Last modified: | 2017-12-20 | | Method: | SOLUTION NMR | | Cite: | A novel lipid transfer protein from the pea Pisum sativum: isolation, recombinant expression, solution structure, antifungal activity, lipid binding, and allergenic properties.
BMC Plant Biol, 16
|
|
2MJ0
 
 | |
2MAL
 
 | |
7RUH
 
 | | Bromodomain-containing protein 4 (BRD4) bromodomain 2 (BD2) complexed with XR844 | | Descriptor: | Bromodomain-containing protein 4, N-{1-[1,1-di(pyridin-2-yl)ethyl]-6-(5-{[(2-fluorophenyl)carbamoyl]amino}-1-methyl-6-oxo-1,6-dihydropyridin-3-yl)-1H-indol-4-yl}-2,2,2-trifluoroethane-1-sulfonamide | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | Deposit date: | 2021-08-17 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bromodomain-containing protein 4 (BRD4) bromodomain 2 (BD2) complexed with XR844
To Be Published
|
|
7RUI
 
 | | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with XR844 | | Descriptor: | Bromodomain-containing protein 4, N-{1-[1,1-di(pyridin-2-yl)ethyl]-6-(5-{[(2-fluorophenyl)carbamoyl]amino}-1-methyl-6-oxo-1,6-dihydropyridin-3-yl)-1H-indol-4-yl}-2,2,2-trifluoroethane-1-sulfonamide | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | Deposit date: | 2021-08-17 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with XR844
To Be Published
|
|
4XQD
 
 | | X-ray structure analysis of xylanase-WT at pH4.0 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XQW
 
 | | X-ray structure analysis of xylanase-N44E with MES at pH6.0 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-20 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XPV
 
 | | Neutron and X-ray structure analysis of xylanase: N44D at pH6 | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-18 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XQ4
 
 | | X-ray structure analysis of xylanase - N44D | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2JWM
 
 | |
2JNI
 
 | | Spatial structure of antimicrobial peptide arenicin-2 in aqueous solution | | Descriptor: | Arenicin-2 | | Authors: | Ovchinnikova, T.V, Shenkarev, Z.O, Nadezhdin, K.D, Balandin, S.V, Zhmak, M.N, Kudelina, I.A, Finkina, E.I, Kokryakov, V.N, Arseniev, A.S. | | Deposit date: | 2007-01-25 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Recombinant expression, synthesis, purification, and solution structure of arenicin
Biochem.Biophys.Res.Commun., 360, 2007
|
|
6P05
 
 | | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with compound 27 | | Descriptor: | Bromodomain-containing protein 4, GLYCEROL, N-{1-[1,1-di(pyridin-2-yl)ethyl]-6-(1-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-3-yl)-1H-indol-4-yl}ethanesulfonamide | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Zhao, J, Gutgesell, L.M, Shen, Z, Dye, K, Dubrovyskyii, O, Zhao, H, Huang, F, Tonetti, D.A, Thatcher, G.R. | | Deposit date: | 2019-05-16 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Novel Pyrrolopyridone Bromodomain and Extra-Terminal Motif (BET) Inhibitors Effective in Endocrine-Resistant ER+ Breast Cancer with Acquired Resistance to Fulvestrant and Palbociclib.
J.Med.Chem., 63, 2020
|
|
3QPS
 
 | | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni | | Descriptor: | CHOLIC ACID, CmeR | | Authors: | Lei, H.T, Routh, M.D, Shen, Z, Su, C.C, Zhang, Q, Yu, E.W. | | Deposit date: | 2011-02-14 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni.
Protein Sci., 20, 2011
|
|
3QQA
 
 | | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni | | Descriptor: | CmeR, TAUROCHOLIC ACID | | Authors: | Lei, H.T, Routh, M.D, Shen, Z, Su, C.-C, Zhang, Q, Yu, E.W. | | Deposit date: | 2011-02-15 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni.
Protein Sci., 20, 2011
|
|
