6JRE
 
 | | Structure of N-terminal domain of Plasmodium vivax p43 (PfNTD) solved by Co-SAD phasing | | Descriptor: | Aminoacyl-tRNA synthetase-interacting multifunctional protein p43, COBALT (II) ION | | Authors: | Manickam, Y, Harlos, K, Sharma, M, Gupta, S, Sharma, A. | | Deposit date: | 2019-04-03 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structures of the two domains that constitute the Plasmodium vivax p43 protein.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
4UAT
 
 | | Crystal structure of CbbY (mutant D10N) from Rhodobacter sphaeroides in complex with Xylulose-(1,5)bisphosphate, crystal form I | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, Protein CbbY, ... | | Authors: | Bracher, A, Sharma, A, Starling-Windhof, A, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2014-08-11 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Degradation of potent Rubisco inhibitor by selective sugar phosphatase.
Nat.Plants, 1, 2015
|
|
4UAR
 
 | | Crystal structure of apo-CbbY from Rhodobacter sphaeroides | | Descriptor: | GLYCEROL, Protein CbbY | | Authors: | Bracher, A, Sharma, A, Starling-Windhof, A, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2014-08-11 | | Release date: | 2014-12-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Degradation of potent Rubisco inhibitor by selective sugar phosphatase.
Nat.Plants, 1, 2015
|
|
4UAV
 
 | | Crystal structure of CbbY (AT3G48420) from Arabidobsis thaliana | | Descriptor: | Haloacid dehalogenase-like hydrolase domain-containing protein At3g48420, MAGNESIUM ION | | Authors: | Bracher, A, Sharma, A, Starling-Windhof, A, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2014-08-11 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Degradation of potent Rubisco inhibitor by selective sugar phosphatase.
Nat.Plants, 1, 2015
|
|
4UAU
 
 | | Crystal structure of CbbY (mutant D10N) from Rhodobacter sphaeroides in complex with Xylulose-(1,5)bisphosphate, crystal form II | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, Protein CbbY, ... | | Authors: | Bracher, A, Sharma, A, Starling-Windhof, A, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2014-08-11 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Degradation of potent Rubisco inhibitor by selective sugar phosphatase.
Nat.Plants, 1, 2015
|
|
3VGJ
 
 | | Crystal of Plasmodium falciparum tyrosyl-tRNA synthetase (PfTyrRS)in complex with adenylate analog | | Descriptor: | ADENOSINE MONOPHOSPHATE, TYROSINE, Tyrosyl-tRNA synthetase, ... | | Authors: | Banday, M.M, Yogavel, M, Bhatt, T.K, Khan, S, Sharma, A, Sharma, A. | | Deposit date: | 2011-08-14 | | Release date: | 2012-07-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.212 Å) | | Cite: | Malaria parasite tyrosyl-tRNA synthetase secretion triggers pro-inflammatory responses.
Nat Commun, 2, 2011
|
|
7X09
 
 | | Homo sapiens Prolyl-tRNA Synthetase (HsPRS) in Complex with inhibitors L95 and Halofuginone | | Descriptor: | 7-bromo-6-chloro-3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, BROMIDE ION, Bifunctional glutamate/proline--tRNA ligase, ... | | Authors: | Manickam, Y, Babbar, P, Pillai, P, Sharma, A. | | Deposit date: | 2022-02-21 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Homo sapiens Prolyl-tRNA synthetase (HsPRS) with double inhibitors (HF and L95)
To Be Published
|
|
7X1O
 
 | |
7VC1
 
 | | Toxoplasma gondii Prolyl-tRNA Synthetase (TgPRS) in Complex with inhibitor L95 and L-proline in space group P21 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Yogavel, M, Malhotra, N, Sharma, A. | | Deposit date: | 2021-09-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Targeting prolyl-tRNA synthetase via a series of ATP-mimetics to accelerate drug discovery against toxoplasmosis.
Plos Pathog., 19, 2023
|
|
7VC3
 
 | | Toxoplasma gondii Prolyl-tRNA Synthetase (TgPRS) in Complex with inhibitor L97 and L-proline at 1.97 A resolution | | Descriptor: | 4-[(3S)-3-cyclopropyl-3-(hydroxymethyl)-2-oxidanylidene-pyrrolidin-1-yl]-N-[[3-fluoranyl-5-(1-methylpyrazol-4-yl)phenyl]methyl]-6-methyl-pyridine-2-carboxamide, CHLORIDE ION, ETHANOLAMINE, ... | | Authors: | Malhotra, N, Yogavel, M, Sharma, A. | | Deposit date: | 2021-09-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Targeting prolyl-tRNA synthetase via a series of ATP-mimetics to accelerate drug discovery against toxoplasmosis.
Plos Pathog., 19, 2023
|
|
7VC2
 
 | | Toxoplasma gondii Prolyl-tRNA Synthetase (TgPRS) in Complex with inhibitor L96 and L-proline in space group P21 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3S)-3-cyano-3-cyclopropyl-2-oxidanylidene-pyrrolidin-1-yl]-N-[[3-fluoranyl-5-(5-methoxypyridin-3-yl)phenyl]methyl]-6-methyl-pyridine-2-carboxamide, CHLORIDE ION, ... | | Authors: | Yogavel, M, Malhotra, N, Sharma, A. | | Deposit date: | 2021-09-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Targeting prolyl-tRNA synthetase via a series of ATP-mimetics to accelerate drug discovery against toxoplasmosis.
Plos Pathog., 19, 2023
|
|
4TWA
 
 | |
5WRU
 
 | |
5X7V
 
 | |
5WRT
 
 | |
5ZKH
 
 | |
7V8K
 
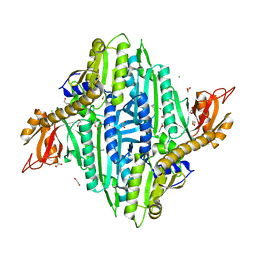 | |
7V8J
 
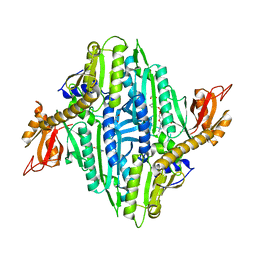 | |
7VC5
 
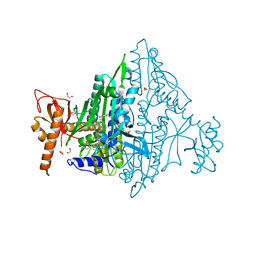 | | Toxoplasma gondii Prolyl-tRNA Synthetase (TgPRS) in Complex with inhibitor T35 and Febrifugine (FF) at 1.61 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, ... | | Authors: | Manickam, Y, Malhotra, N, Sharma, A. | | Deposit date: | 2021-09-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.612 Å) | | Cite: | TgPRS with double inhibitors
To Be Published
|
|
5ZDL
 
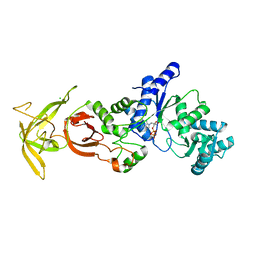 | | Crystal Structure Analysis of TtQRS in co-crystallised with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Glutamine--tRNA ligase | | Authors: | Mutharasappan, N, Jain, V, Sharma, A, Manickam, Y, Jeyaraman, J. | | Deposit date: | 2018-02-23 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional analysis of Glutaminyl-tRNA synthetase (TtGlnRS) from Thermus thermophilus HB8 and its complexes
Int. J. Biol. Macromol., 120, 2018
|
|
7V9D
 
 | | Plasmodium falciparum Prolyl-tRNA Synthetase (PfPRS) in Complex with inhibitor L95 and azetidine | | Descriptor: | (2S)-azetidine-2-carboxylic acid, 1,2-ETHANEDIOL, 1,4-BUTANEDIOL, ... | | Authors: | Manickam, Y, Malhotra, N, Sharma, A. | | Deposit date: | 2021-08-24 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | Plasmodium falciparum Prolyl-tRNA Synthetase (PfPRS) in Complex with inhibitor L95 and azetidine
To Be Published
|
|
6A88
 
 | | Crystal Structure of T. gondii prolyl tRNA synthetase with Febrifugine and ATP Analog | | Descriptor: | 3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Kumari, S, Mishra, S, Yogavel, M, Sharma, A. | | Deposit date: | 2018-07-06 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Conformational heterogeneity in apo and drug-bound structures of Toxoplasma gondii prolyl-tRNA synthetase.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
5X6N
 
 | | Structure of P. Knowlesi DBL Domain Capable of binding Human Duffy Antigen | | Descriptor: | Duffy binding protein, SULFATE ION, octyl beta-D-glucopyranoside | | Authors: | Singh, S.K, Hora, R, Belrhali, H, Chitnis, C, Sharma, A. | | Deposit date: | 2017-02-22 | | Release date: | 2017-03-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for Duffy recognition by the malaria parasite Duffy-binding-like domain
Nature, 439, 2006
|
|
5XIF
 
 | | Crystal Structure of Prolyl-tRNA Synthetase (PRS) from Toxoplasma gondii | | Descriptor: | CHLORIDE ION, Prolyl-tRNA synthetase (ProRS) | | Authors: | Jain, V, Manickam, Y, Sharma, A. | | Deposit date: | 2017-04-26 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Targeting Prolyl-tRNA Synthetase to Accelerate Drug Discovery against Malaria, Leishmaniasis, Toxoplasmosis, Cryptosporidiosis, and Coccidiosis
Structure, 25, 2017
|
|
5XIQ
 
 | | Crystal Structure of Toxoplasma gondii Prolyl-tRNA Synthetase (TgPRS) in complex with Halofuginone | | Descriptor: | 7-bromo-6-chloro-3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Jain, V, Manickam, Y, Sharma, A. | | Deposit date: | 2017-04-26 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Targeting Prolyl-tRNA Synthetase to Accelerate Drug Discovery against Malaria, Leishmaniasis, Toxoplasmosis, Cryptosporidiosis, and Coccidiosis
Structure, 25, 2017
|
|
